Search Count: 49
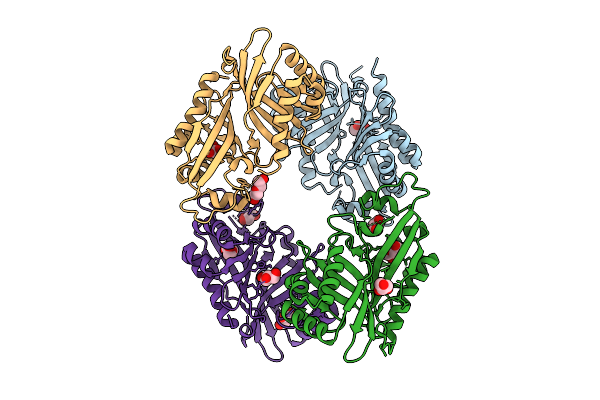 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
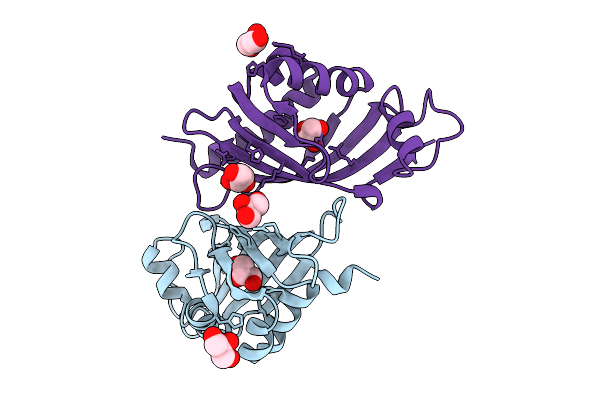 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
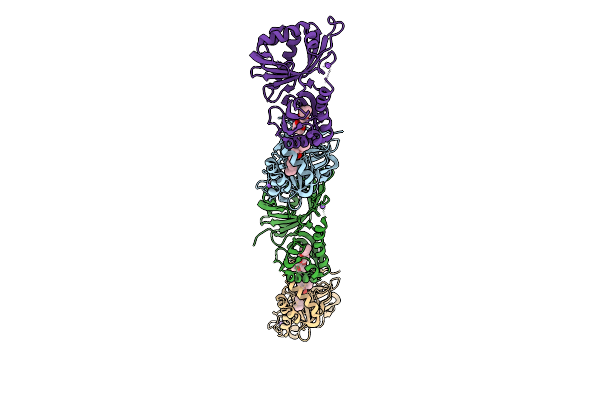 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-05-28 Classification: IMMUNE SYSTEM Ligands: GOL, EDO, PEG, K, CL |
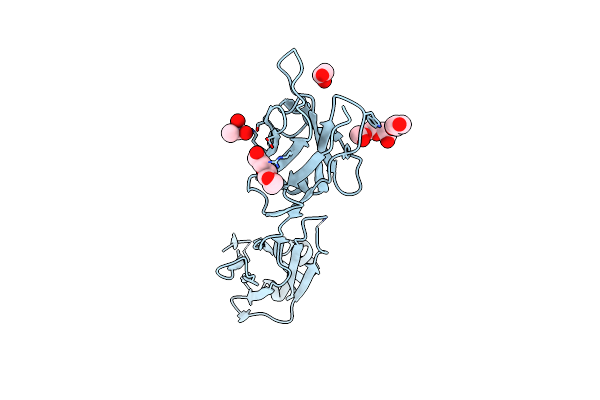 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-28 Classification: IMMUNE SYSTEM Ligands: CAD, EDO, PEG, PGE |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 100 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Dark State Sfx Structure Of Hen Egg White Lysozyme Microcrystals Immobilized With A Mn Carbonyl Complex At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 10 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation With 40 Microjoules Laser Intensity At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Dihydrofolate Reductase-Like Enzyme From Leptospira Interrogans (Selenomethionine Derivative)
Organism: Leptospira interrogans serovar pomona
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: 1PE, NAP, SO4 |
 |
Organism: Leptospira interrogans serovar pomona
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: 1PE, NAP, SO4, ACT |
 |
Dihydrofolate Reductase-Like Enzyme From Leptospira Interrogans With Additional Nadp+
Organism: Leptospira interrogans serovar pomona
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: 1PE, NAP, SO4, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-03-13 Classification: CYTOKINE Ligands: SO4, EDO, IPA |
 |
Crystal Structure Of An Antibody Light Chain Tetramer With 3D Domain Swapping
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: IMMUNE SYSTEM |
 |
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Globiformis Anaerobically Reduced By Phenylethylamine At Pd 9.0
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.09 Å, 1.67 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
 |
Neutron Structure Of [Nife]-Hydrogenase From D. Vulgaris Miyazaki F In Its Oxidized State
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.04 Å, 2.20 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NFU, MPD, OH, MG, CL, SF4, F3S |
 |
High Resolution Crystal Structure Of Human Macrophage Migration Inhibitory Factor In Complex With Methotrexate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2023-05-24 Classification: CYTOKINE/INHIBITOR Ligands: SO4, EDO, IPA, MT1 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc, D122G/H130Y/T267C Mutant, Hydroxylamine-Treated
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2023-03-29 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Structure Of 6-Aminohexanoate-Oligomer Hydrolase Nylc Precursor, H130Y/N266A/T267A Mutant
Organism: Arthrobacter
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: GOL, SO4, NA |

