Search Count: 19
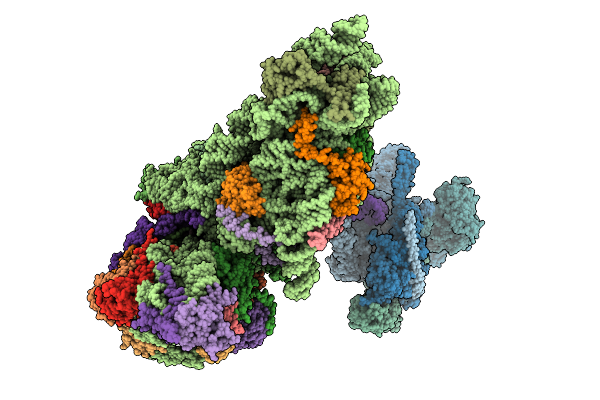 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
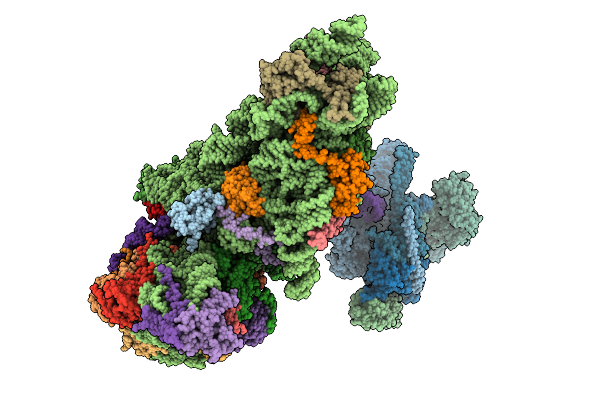 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
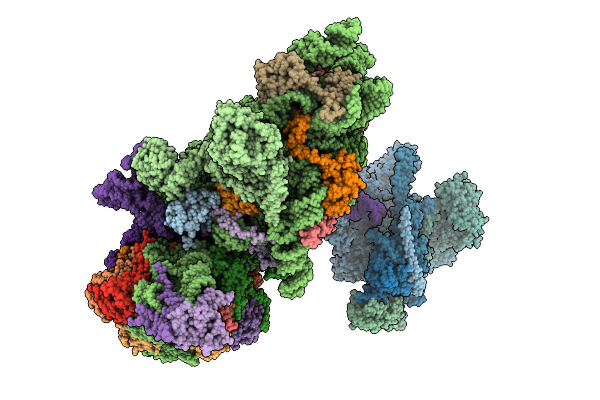 |
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
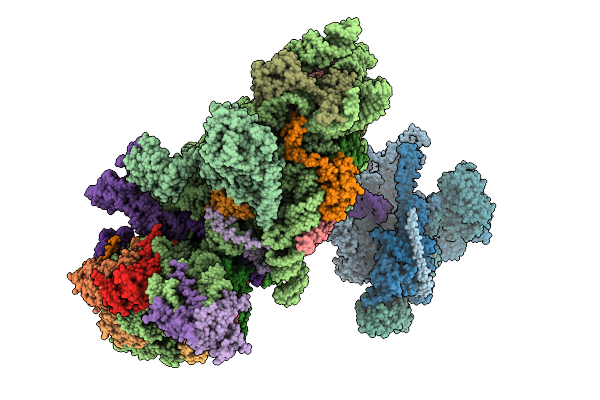 |
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
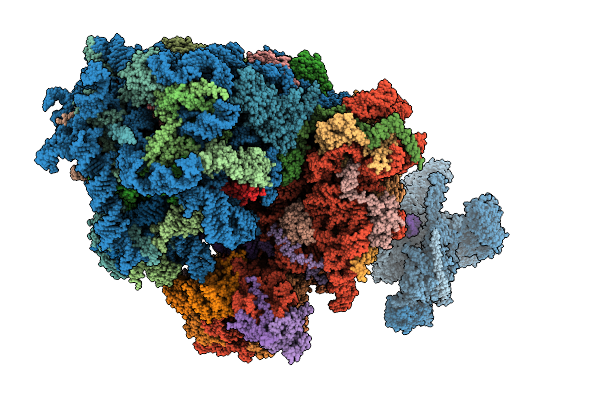 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
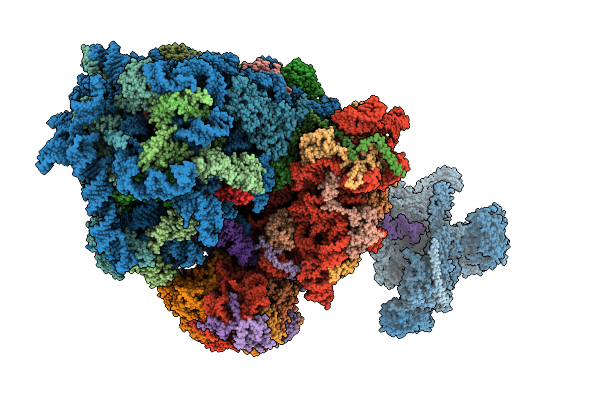 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
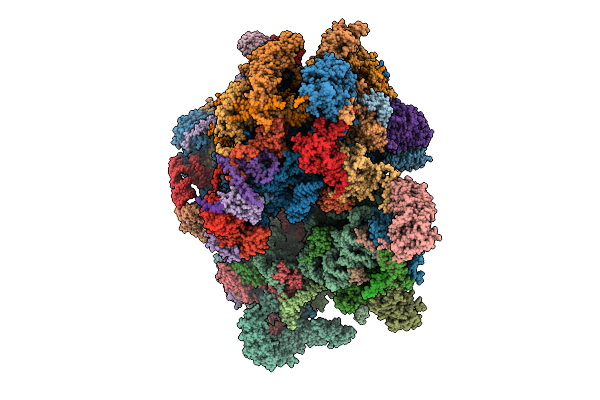 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, K, G34, VAL, NAD, SPD, ZN, FES, ATP, GDP |
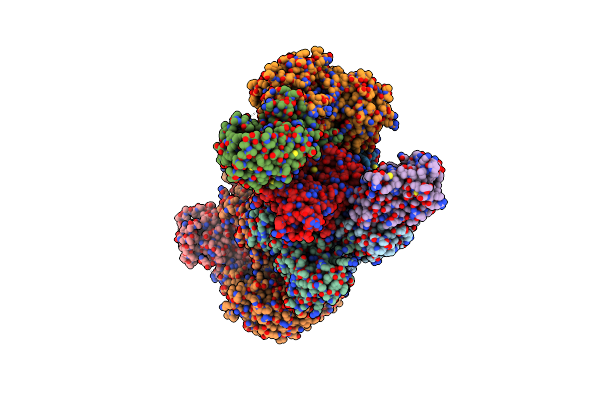 |
Organism: Homo sapiens, Sandfly fever sicilian virus
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: TRANSLATION |
 |
Organism: Homo sapiens, Sandfly fever sicilian virus
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: TRANSLATION |
 |
Organism: Homo sapiens, Sandfly fever sicilian virus
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: TRANSLATION |
 |
Organism: Homo sapiens, Sandfly fever sicilian virus
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: TRANSLATION |
 |
Crystal Structure Of The Human Eif4A1-Atp Analog-Roca-Polypurine Rna Complex
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: TRANSLATION/RNA Ligands: ANP, MG, RCG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-12-05 Classification: TRANSFERASE |
 |
Crystal Structure Of The Human Cap-Specific Adenosine Methyltransferase Bound To Sah
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, EDO |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah And M7G-Capped Rna
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-05 Classification: TRANSFERASE Ligands: SAH, EDO, M7G |
 |
Crystal Structure Of The Zebrafish Cap-Specific Adenosine Methyltransferase Bound To Sah And M7G-Capped Rna
Organism: Danio rerio, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-05 Classification: TRANSFERASE/RNA Ligands: SAH, M7G, EDO, B3P |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-04-25 Classification: RNA BINDING PROTEIN Ligands: GOL |

