Search Count: 13
 |
Crystal Structure Of The Glua2 Ligand Binding Core (S1S2J) In Complex With Fluorophore-Ligand Conjugate
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-07-02 Classification: SIGNALING PROTEIN Ligands: A1L33, SO4, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2018-11-21 Classification: signaling protein/antagonist Ligands: CF7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2018-03-21 Classification: transcription/agonist Ligands: MPD, E3S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-03-21 Classification: transcription/agonist Ligands: MPD, E3V |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-01-03 Classification: SIGNALING PROTEIN Ligands: CFG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-02 Classification: LIGASE/LIGASE inhibitor Ligands: CA, CL |
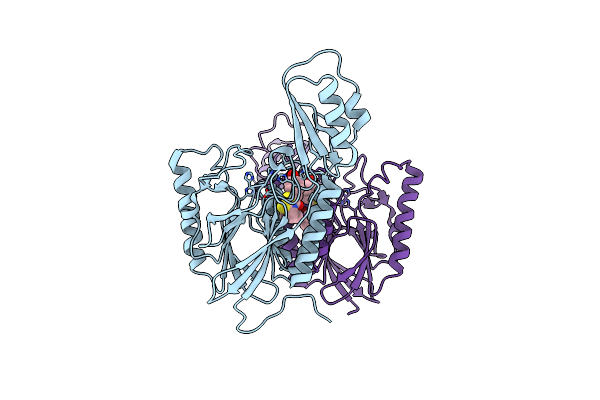 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-15 Classification: DNA BINDING PROTEIN/inhibitor Ligands: 2Q0, MN |
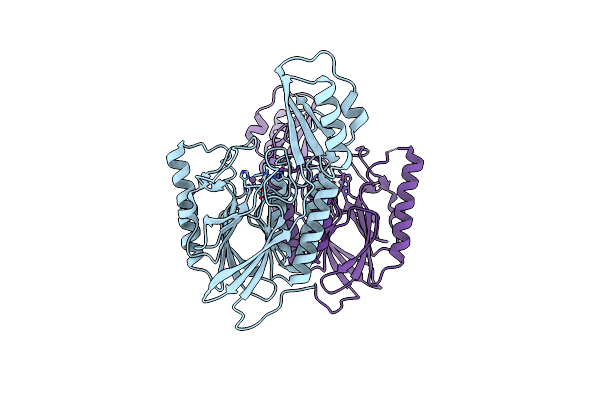 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-08 Classification: HYDROLASE Ligands: MN |
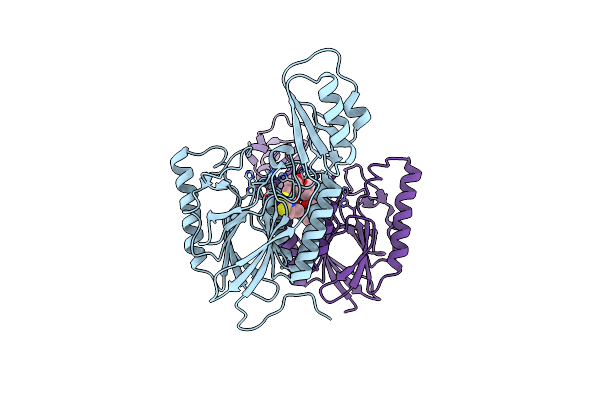 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PW |
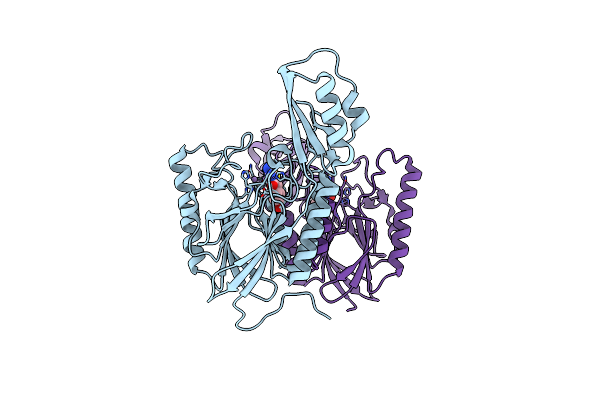 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PK |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PV |
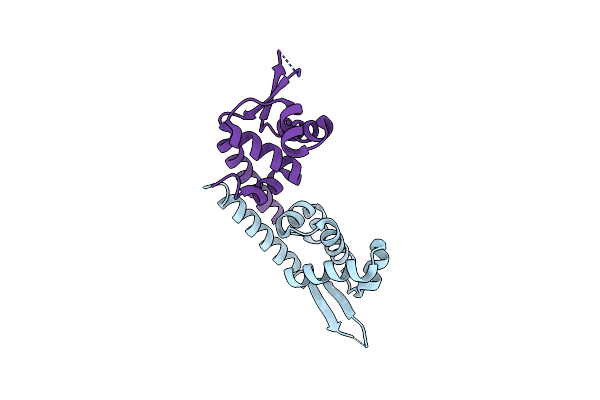 |
Crystal Structure Of St1889 Protein From Thermoacidophilic Archaeon Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-09-05 Classification: TRANSCRIPTION |
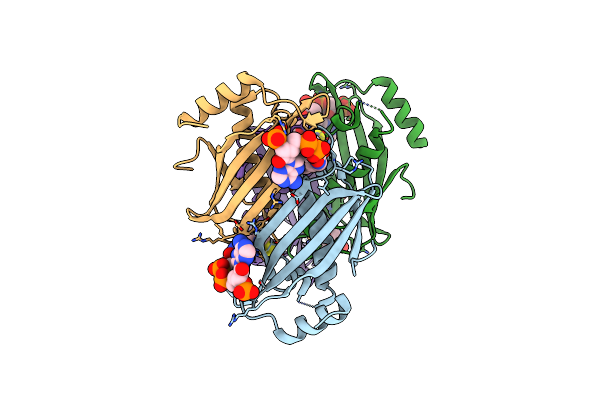 |
Crystal Structure Of Thioesterase Complexed With Coenzyme A And Zn From Thermus Thermophilus Hb8
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-01-06 Classification: HYDROLASE Ligands: ZN, COA |

