Search Count: 300
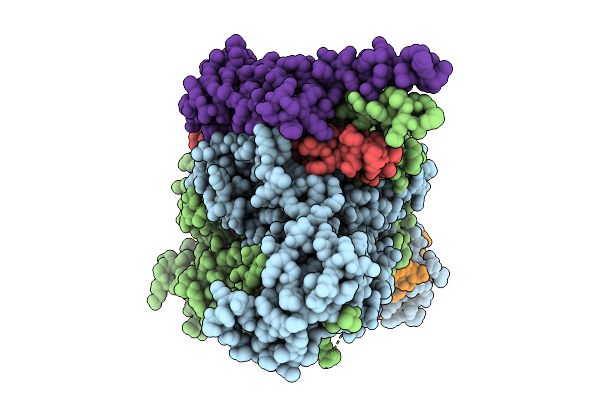 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
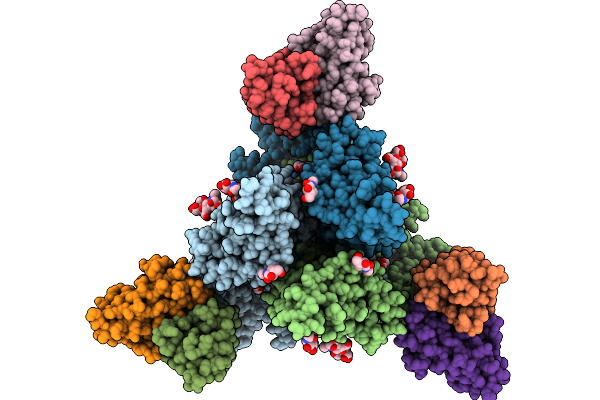 |
Cryo-Em Structure Of Neutralizing Murine Antibody Ws.Hsv-1.24 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P.531E
Organism: Human alphaherpesvirus 1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
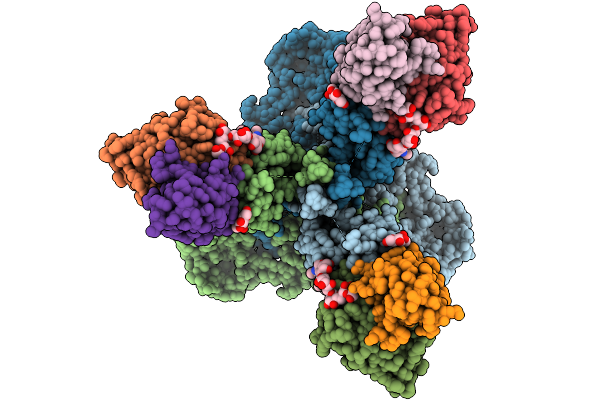 |
Cryo-Em Structure Of Neutralizing Human Antibody D48 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P.531E.Ds
Organism: Human alphaherpesvirus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
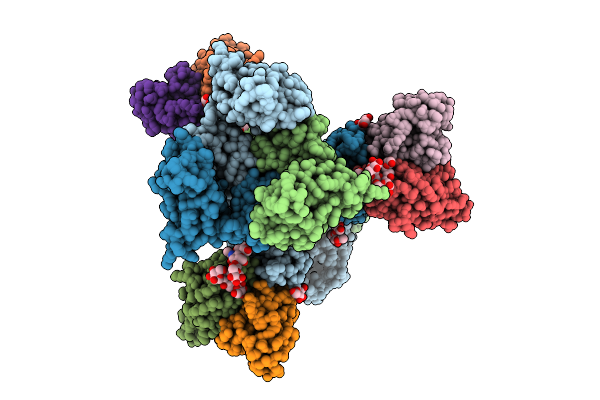 |
Cryo-Em Structure Of Neutralizing Human Antibody D48 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P
Organism: Human alphaherpesvirus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
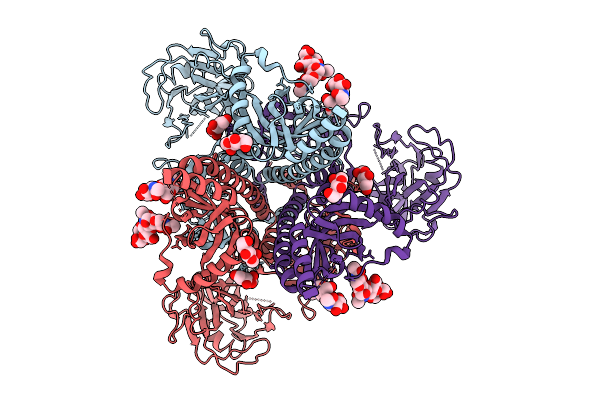 |
Cryo-Em Structure Of Gb-Ecto.516P.531E.Ds, A Prefusion-Stabilized Hsv-1 Glycoprotein B Extracellular Domain
Organism: Human alphaherpesvirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
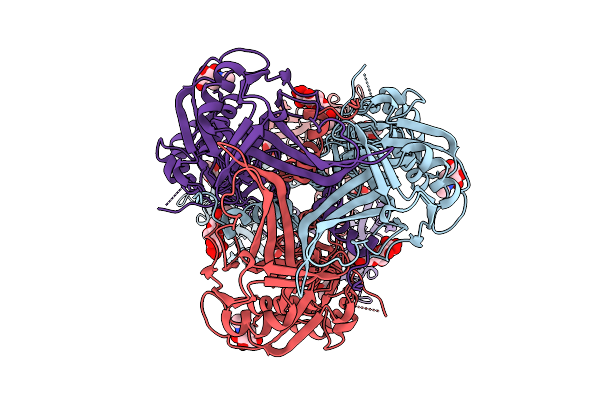 |
Cryo-Em Structure Of Gb-Ecto.516P, An Hsv-1 Glycoprotein B Extracellular Domain
Organism: Human alphaherpesvirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
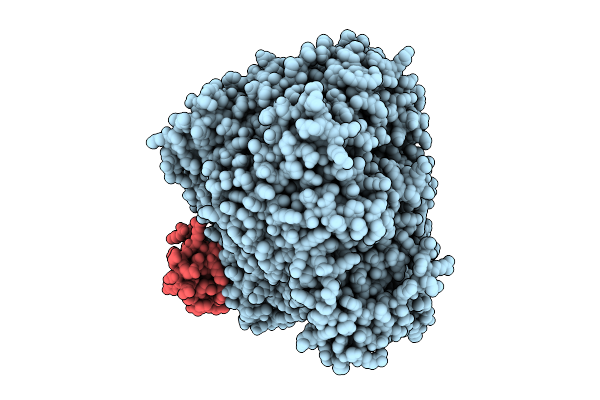 |
Organism: Measles virus strain ichinose-b95a
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSCRIPTION Ligands: ZN |
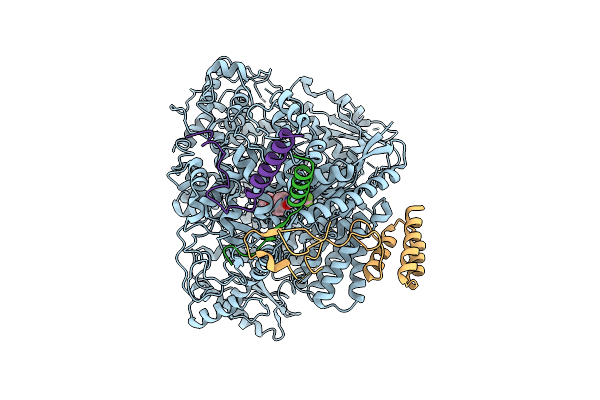 |
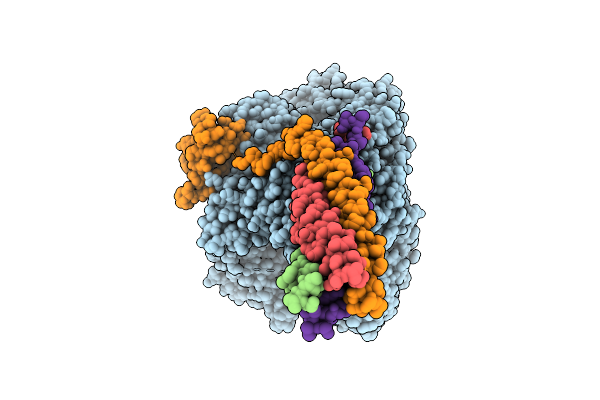
Organism: Measles virus strain ichinose-b95a
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSCRIPTION Ligands: ZN, A1EF9 |
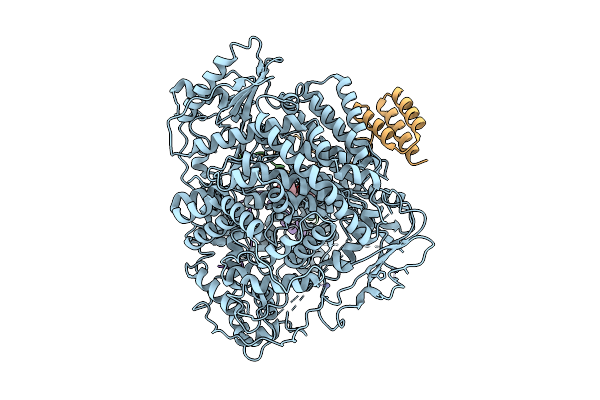 |
Organism: Measles virus strain ichinose-b95a
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSCRIPTION Ligands: ZN, A1EGA |
 |
Organism: Henipavirus nipahense
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSCRIPTION Ligands: A1EF9, ZN |
 |
Organism: Sfts virus js4, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Sfts virus hb29, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
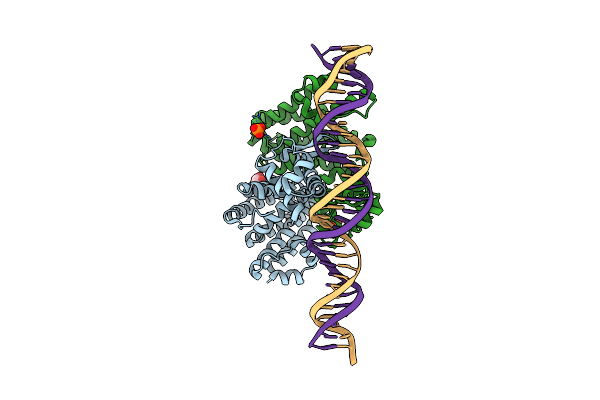
Organism: Severe fever with thrombocytopenia syndrome virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
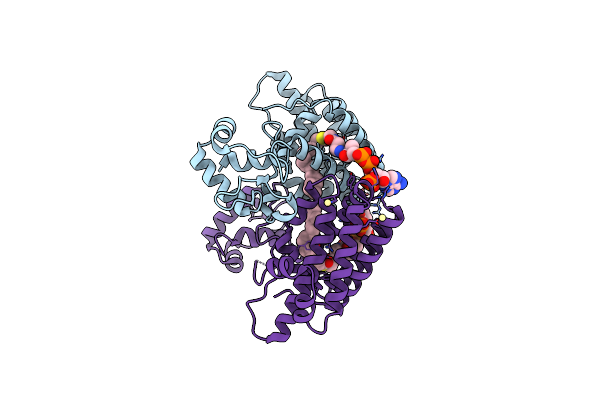
Vcfadrqm, Genetically Engineered Mutants Of Vibrio Cholerae Fadr, In Complex With Dna
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-05 Classification: DNA BINDING PROTEIN Ligands: GOL, PO4 |
 |
Vcfadrqm, Mutant Protein Of Fatty Acid Responsive Transcription Factor From Vibrio Cholerae, In Complex With Oleoyl-Coa
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2025-02-05 Classification: DNA Ligands: 3VV, CD |
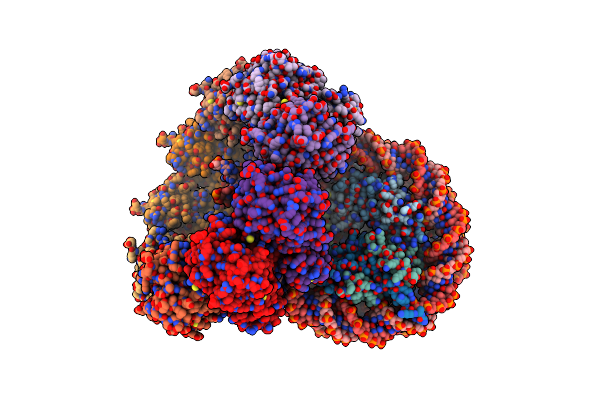 |
The Cryo-Em Structure Of The Nonameric Rad51 Ring Bound To The Nucleosome With The Linker Dna Binding
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DNA BINDING PROTEIN/DNA |
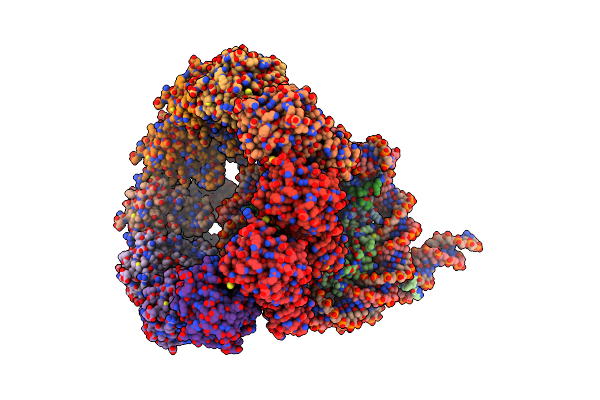 |
The Cryo-Em Structure Of The Decameric Rad51 Ring Bound To The Nucleosome Without The Linker Dna Binding
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DNA BINDING PROTEIN/DNA |
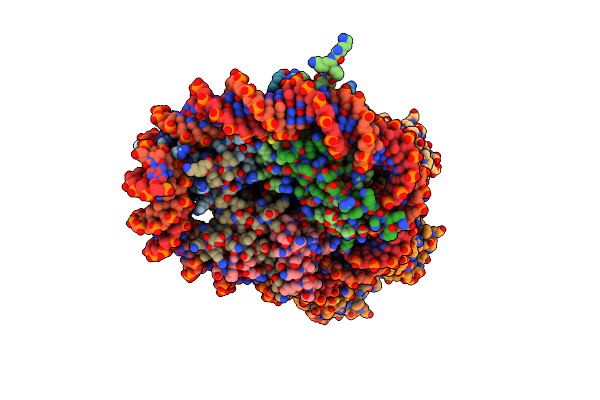 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DNA BINDING PROTEIN/DNA |
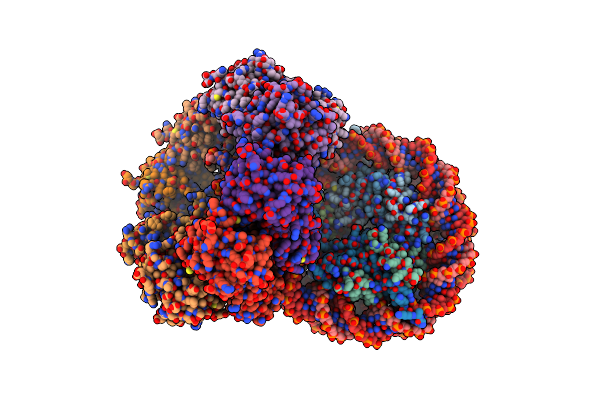 |
The Cryo-Em Structure Of The Octameric Rad51 Ring Bound To The Nucleosome With The Linker Dna Binding
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DNA BINDING PROTEIN/DNA |
 |
The Cryo-Em Structure Of The Decameric Rad51 Ring Bound To The Nucleosome With The Linker Dna Binding
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-27 Classification: DNA BINDING PROTEIN/DNA |

