Search Count: 781
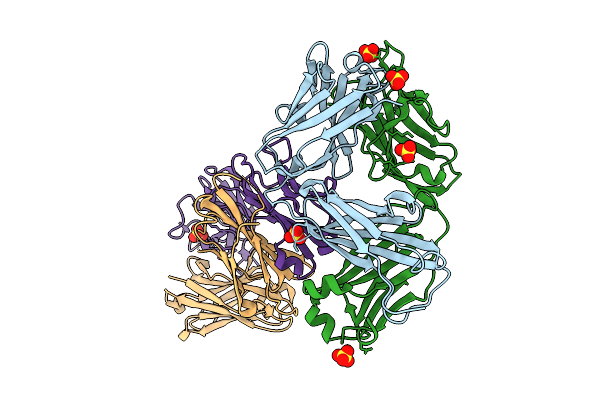 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM Ligands: SO4 |
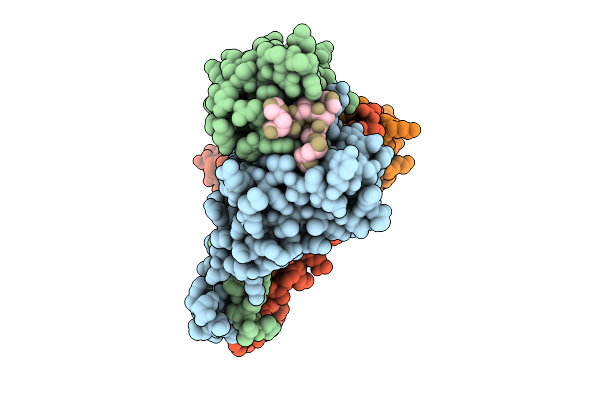 |
Organism: Mus musculus, Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
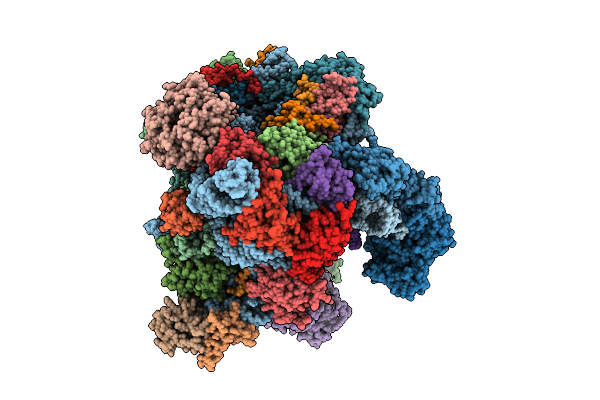 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
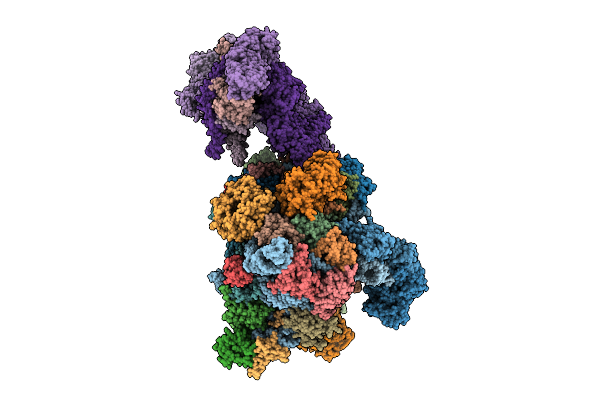 |
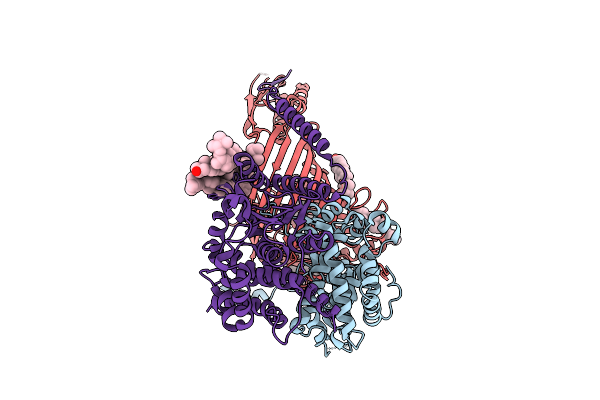
Late-Stage 48S Initiation Complex With Eif3 (Ls48S-Eif3 Ic) Guided By The Trans-Rna
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
 |
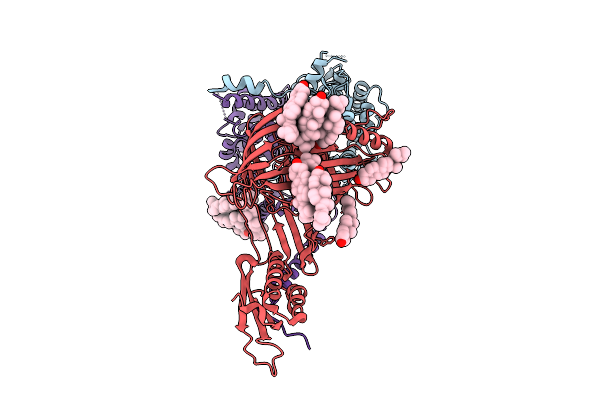
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
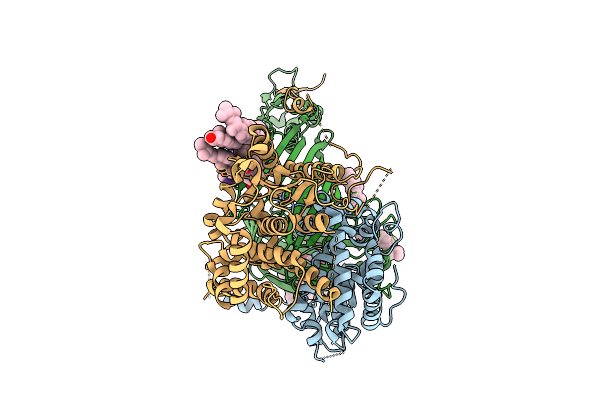
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
 |
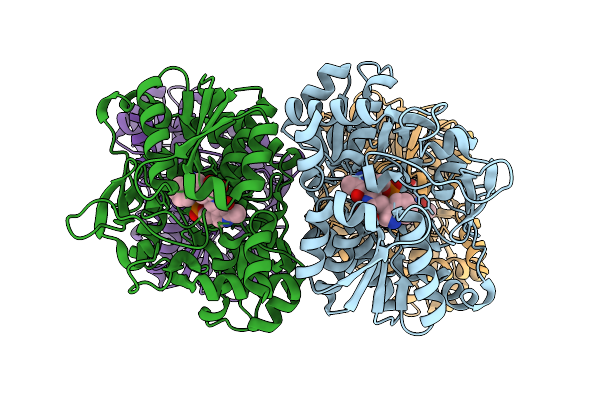
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
 |
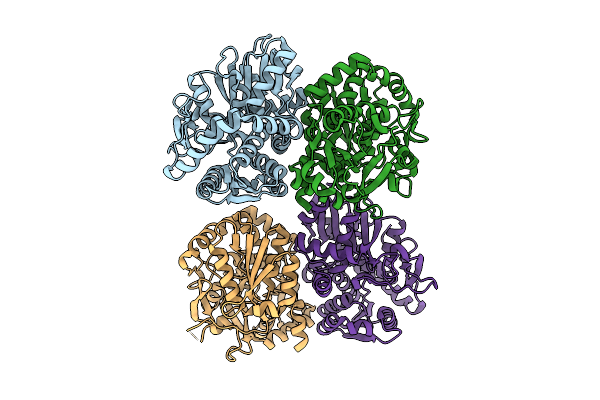
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BIOSYNTHETIC PROTEIN Ligands: VB4 |
 |
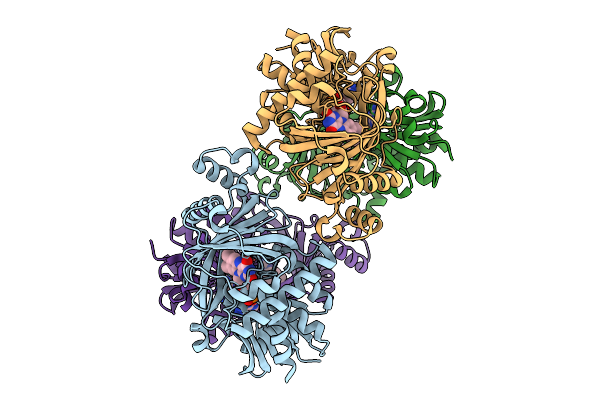
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Vanrija humicola
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: OXIDOREDUCTASE Ligands: FAD, F |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MOTOR PROTEIN |
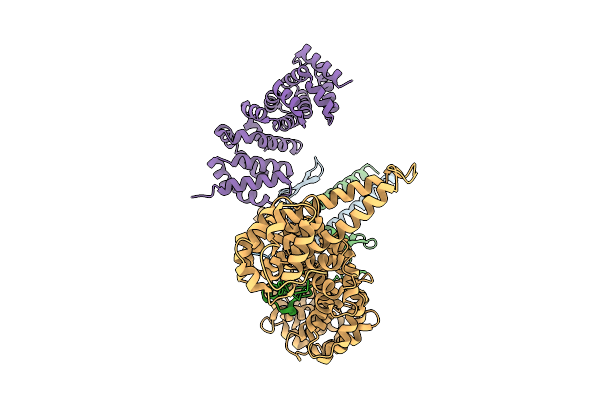 |
Cryo-Em Structure Of The Kinesin-2 Tail Domain In Complex With Kap3 And Apc
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MOTOR PROTEIN |
 |
Organism: Aquimarina spongiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN Ligands: UNL, CL, SO4 |
 |
Organism: Aquimarina spongiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FPP, CA, SO4, CL, A1MAK |
 |
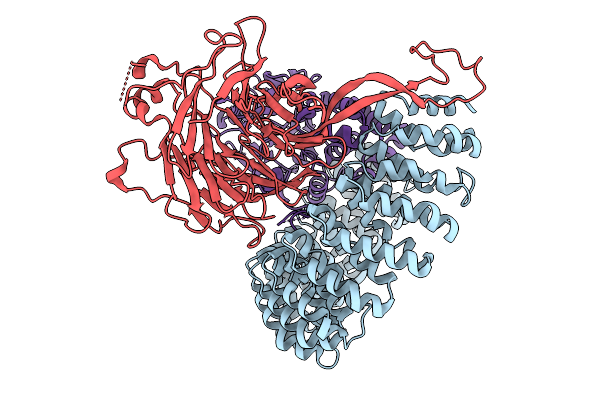
Crystal Structure Of Asdms D333N Mutant In Complex With Farnesyl Pyrophosphate
Organism: Aquimarina spongiae
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FPP, CL, SO4, GOL, MES |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: ONCOPROTEIN |
 |
Cryo-Em Structure Of Da03E17 Fab In Complex With Influenza Virus Neuraminidase From A/California/07/2009 (H1N1)
Organism: Influenza a virus (a/california/07/2009(h1n1)), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, CA |
 |
Cryo-Em Structure Of Da03E17 Fab In Complex With Influenza Virus Neuraminidase From A/Kansas/14/2017 (H3N2)
Organism: Influenza a virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: CA, NAG |
 |
Cryo-Em Structure Of Influenza Virus Neuraminidase From A/Kansas/14/2017 (H3N2)
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: CA, NAG |
 |
Cryo-Em Structure Of Da03E17 Fab In Complex With Influenza Virus Neuraminidase From B/Colorado/06/2017
Organism: Homo sapiens, Influenza b virus (b/colorado/06/2017)
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: CA |

