Search Count: 299
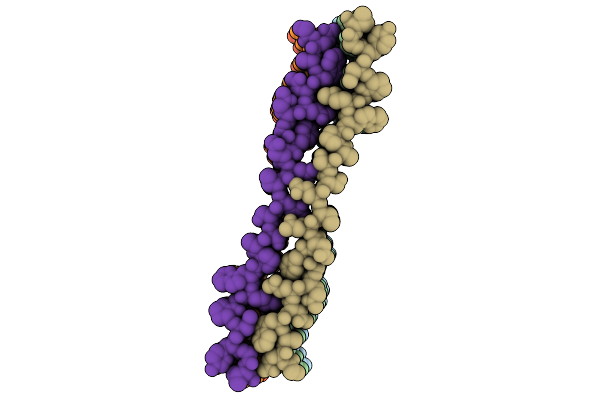 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
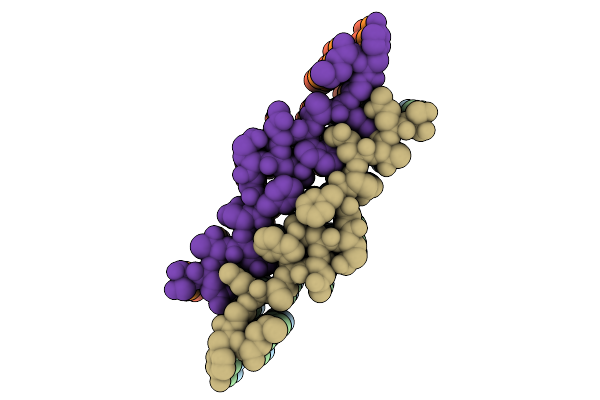 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
 |
Structure Of A De Novo Designed Interleukin-21 Mimetic Complex With Il-21R And Il-2Rg
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |
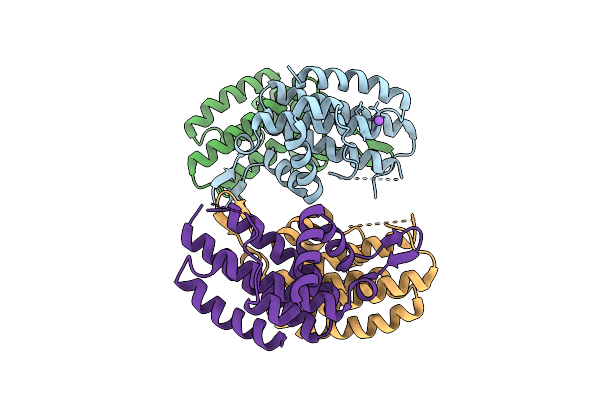 |
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: RNA BINDING PROTEIN |
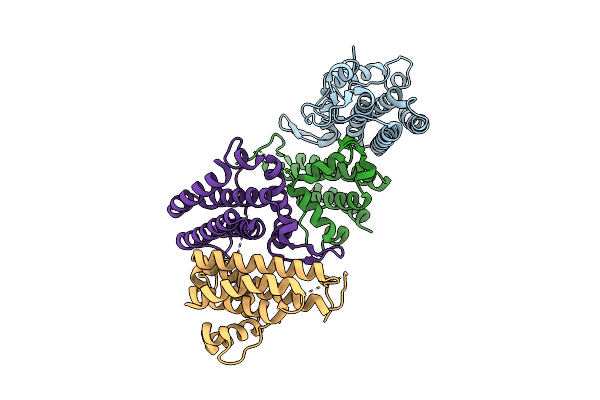 |
Crystal Structure Of The Hepn Family Member Abiv, An Rnase In A Two-Component Antiphage System In Lactococcus Lactis
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: RNA BINDING PROTEIN |
 |
Organism: Lactococcus cremoris
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: RNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-3318
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTM |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With Inhibitor Avi-4692
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: A1BVT, A1BTO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro)In Complex With Inhibitor Avi-4516
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1BTP, EDO |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) Variant Q192T In Complex With Inhibitor Avi-4303
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, A1BTN, CL |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-06-04 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-06-04 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: SIGNALING PROTEIN |
 |
F1 Domain Of Non-Catalytic Site Depleted And Epsilon C-Terminal Domain Deleted Fof1-Atpase From Bacillus Ps3,Under Atp Saturated Condition
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MOTOR PROTEIN Ligands: MG, ADP, ATP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2025-03-19 Classification: TRANSCRIPTION Ligands: A1AAL, EDO |
 |
F1 Domain Of Non-Catalytic Site Depleted And Epsilon C-Terminal Domain Deleted Fof1-Atpase From Bacillus Ps3,Nucleotide Depleted Condition
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-12-18 Classification: TRANSCRIPTION Ligands: YPB, EDO |
 |
Organism: Mus musculus, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: LIPID TRANSPORT Ligands: Y01, LBN |
 |
Organism: Mus musculus, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: LIPID TRANSPORT Ligands: LBN, Y01 |

