Search Count: 22
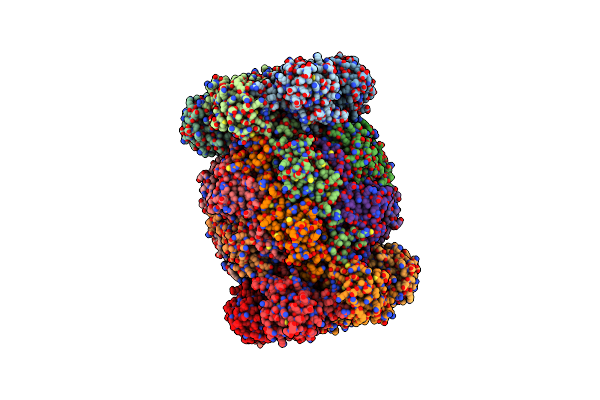 |
Cryoem Structure Of Recombinant Trypanosoma Cruzi Apo Proteasome 20S Subunit
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Resolution:2.25 Å Release Date: 2024-12-25 Classification: UNKNOWN FUNCTION |
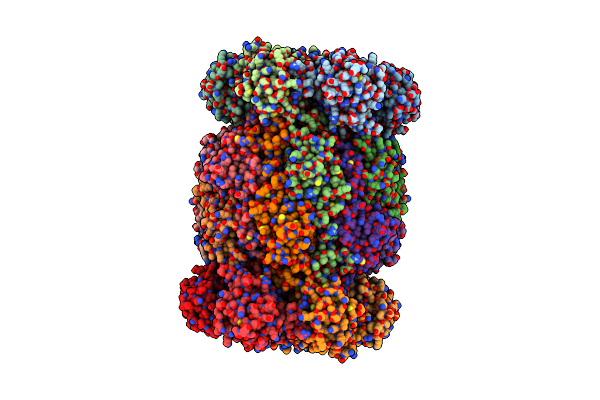 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Resolution:2.31 Å Release Date: 2024-12-25 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, GOL |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine And An Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, C6I |
 |
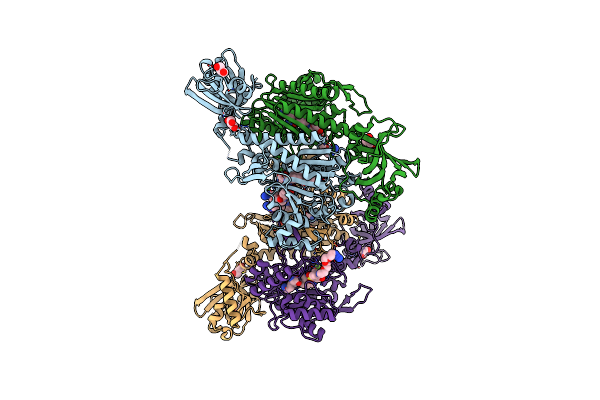
Crystal Structure Of Lysyl-Trna Synthetase From Mycobacterium Tuberculosis Complexed With L-Lysine And Inhibitor
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-10-19 Classification: LIGASE Ligands: LYS, DD0 |
 |
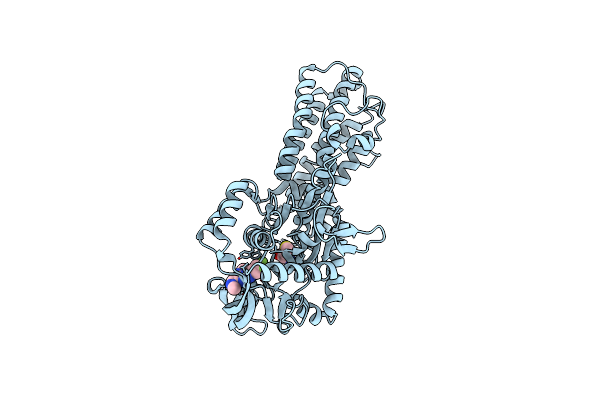
Crystal Structure Of Cryptosporidium Parvum -Plasmodium Falciparum Mutant Lysyl Trna Synthetase In Complex With Inhibitor
Organism: Cryptosporidium parvum iowa ii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: LIGASE Ligands: LYS, JE5, GOL |
 |
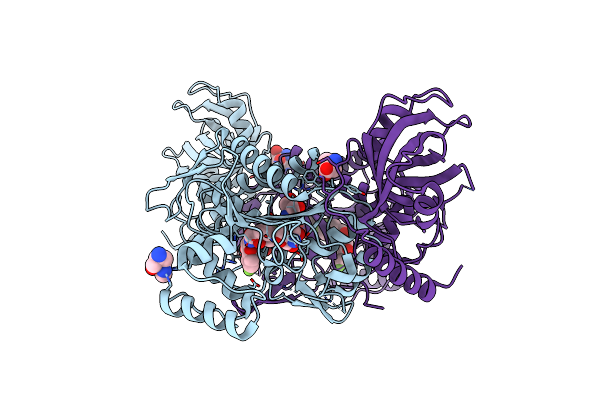
Leishmania Major Methionyl-Trna Synthetase In Complex With An Allosteric Inhibitor
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-08-05 Classification: TRANSLATION Ligands: LWN, MET |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Bound To A Difluoro Cyclohexyl Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYB, LYS, PEG, HIS, TRS |
 |
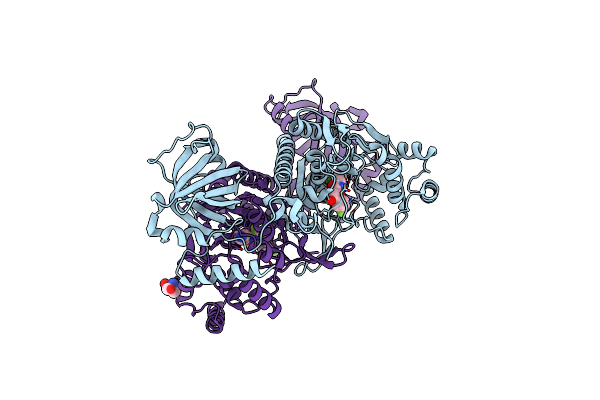
Crystal Structure Of Lysyl-Trna Synthetase From Plasmodium Falciparum Complexed With A Chromone Ligand
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: FYE |
 |
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine And A Difluoro Cyclohexyl Chromone Ligand
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2019-04-03 Classification: TRANSFERASE Ligands: LYS, FYB, TRS |
 |
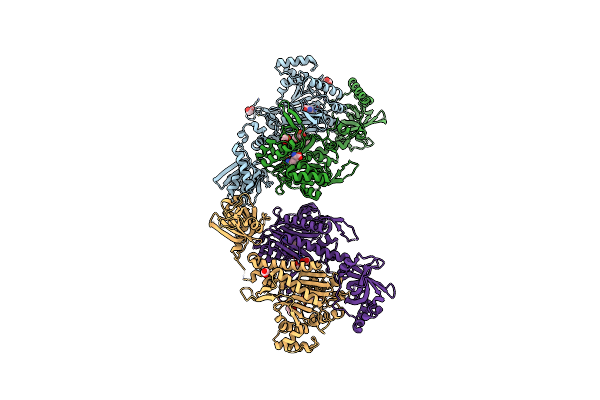
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-03-13 Classification: LIGASE/INHIBITOR Ligands: 9X0, CO, FMT, MLA, LYS |
 |
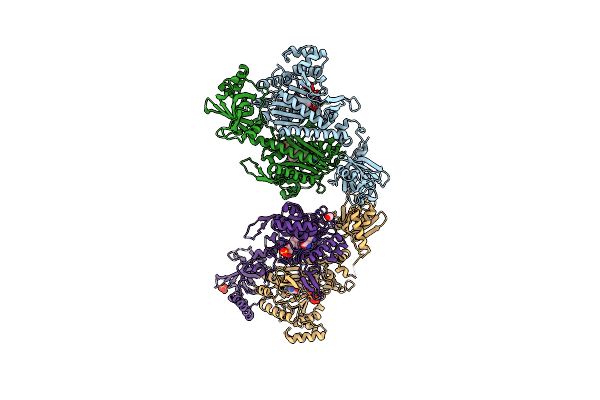
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: LYS, EDO, GOL |
 |
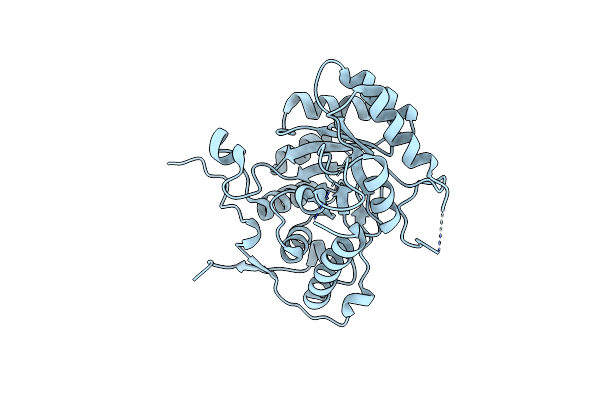
Crystal Structure Of Lysyl-Trna Synthetase From Cryptosporidium Parvum Complexed With L-Lysine And Cladosporin
Organism: Cryptosporidium parvum (strain iowa ii)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: ligase/ligase inhibitor Ligands: LYS, KRS, SO4, EDO |
 |
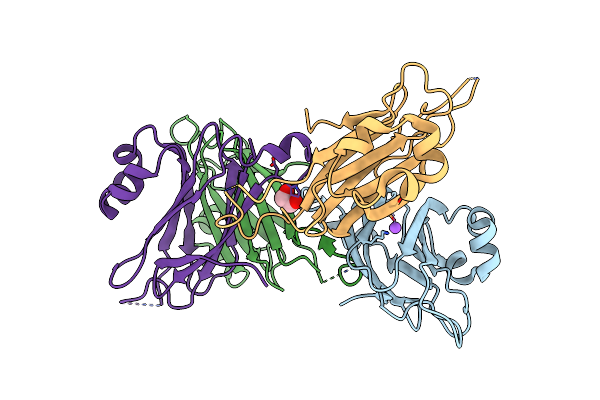
Crystal Structure Of Pseudomonas Aeruginosa Inosine 5'-Monophosphate Dehydrogenase
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-01-16 Classification: OXIDOREDUCTASE Ligands: CL |
 |
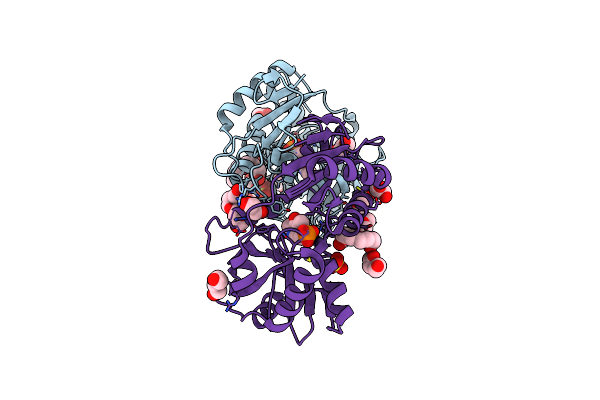
The Structure Of Serratia Marcescens Lip, A Membrane Bound Component Of The Type Vi Secretion System.
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-10-05 Classification: MEMBRANE PROTEIN |
 |
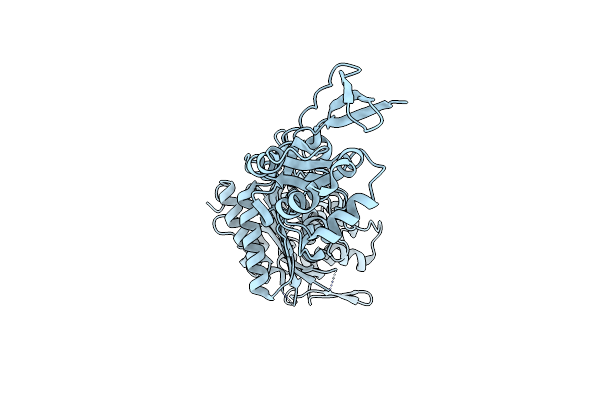
Crystal Structure Of 4-Amino-4-Deoxychorismate Lyase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-01-19 Classification: LYASE Ligands: PLP, PG4, CL, SO4, EDO, PEG, GOL |
 |
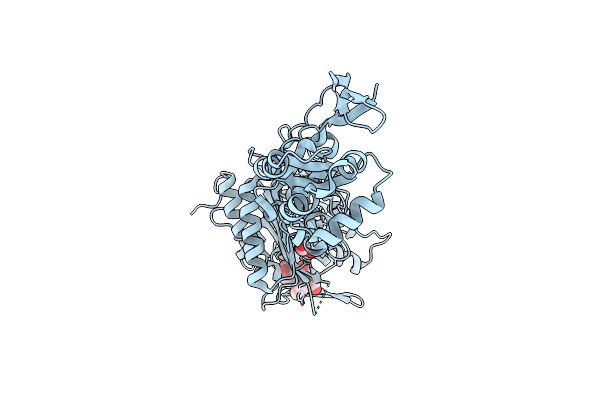
Crystal Structure Of Penicillin-Binding Protein 3 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2010-11-10 Classification: PENICILLIN-BINDING PROTEIN Ligands: CL |
 |
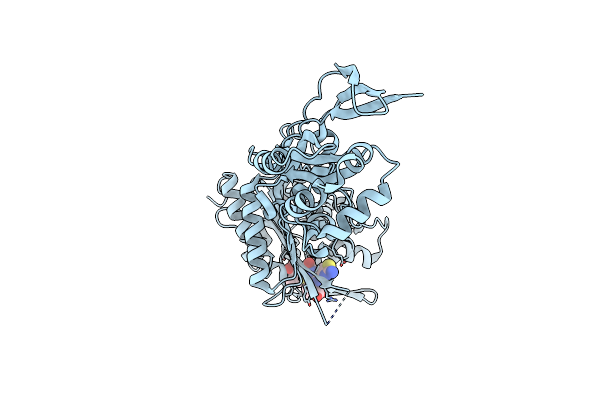
Crystal Structure Of Penicillin-Binding Protein 3 From Pseudomonas Aeruginosa In Complex With Carbenicillin
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-11-10 Classification: PENICILLIN-BINDING PROTEIN/ANTIBIOTIC Ligands: CL, CB9, GOL |
 |
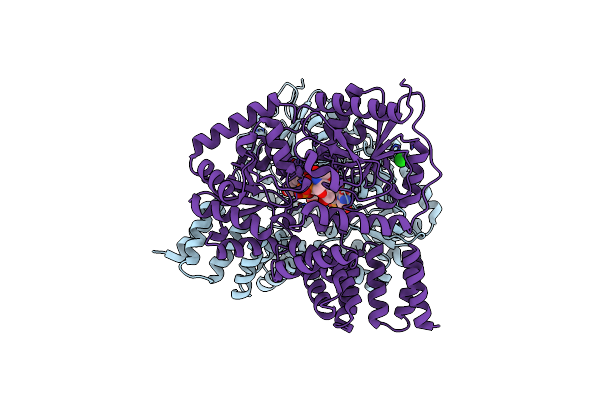
Crystal Structure Of Penicillin-Binding Protein 3 From Pseudomonas Aeruginosa In Complex With Ceftazidime
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2010-11-10 Classification: PENICILLIN-BINDING PROTEIN/ANTIBIOTIC Ligands: CTJ |
 |
Xanthomonas Campestris Putative Ogt (Xcc0866), Complex With Udp- Glcnac Phosphonate Analogue
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-11-25 Classification: TRANSFERASE Ligands: UDM, MN, CL |

