Search Count: 23
 |
Crystal Structure Of Nitrite Bound Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, PG4, NO2 |
 |
Crystal Structure Of As Isolated Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia sp. 5_2_56faa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, CL, PG4 |
 |
Crystal Structure Of As Isolated Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC |
 |
Crystal Structure Of Nitrite Bound Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, NO2 |
 |
Crystal Structure Of As Isolated Y323E Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, GOL |
 |
Crystal Structure Of Nitrite Bound Y323E Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, NO2, GOL |
 |
Crystal Structure Of As Isolated Y323F Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, GOL |
 |
Crystal Structure Of Nitrite Bound Y323F Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, NO2, HEC |
 |
Oxidised Copper Nitrite Reductase From Achromobacter Cycloclastes Determined By Serial Femtosecond Rotation Crystallography
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
 |
Nitrite-Bound Copper Nitrite Reductase From Achromobacter Cycloclastes Determined By Serial Femtosecond Rotation Crystallography
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, NO2, SO4, MLI |
 |
Reduced Copper Nitrite Reductase From Achromobacter Cycloclastes Determined By Serial Femtosecond Rotation Crystallography
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, MLI |
 |
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, NO2, MLI |
 |
Neutron Crystal Structure For Copper Nitrite Reductase From Achromobacter Cycloclastes At 1.8 A Resolution
Organism: Achromobacter cycloclastes
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, NO2, MLI |
 |
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, NO2, MLI |
 |
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: CU, NO2, MLI |
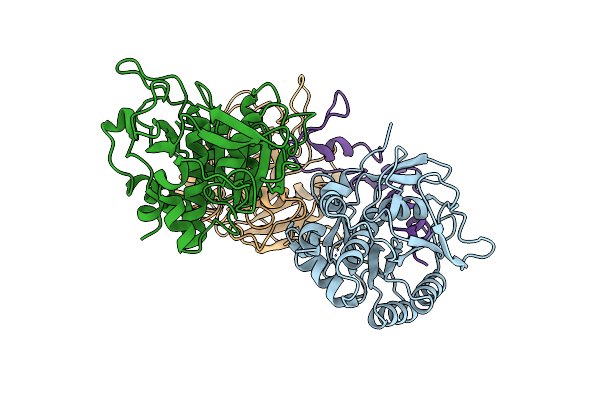 |
Structural Basis For Dual-Inhibition Mechanism Of A Non-Classical Kazal-Type Serine Protease Inhibitor From Horseshoe Crab In Complex With Subtilisin
Organism: Bacillus licheniformis, Carcinoscorpius rotundicauda
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-06-01 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Structural Basis For Reversible And Irreversible Inhibition Of Human Cathepsin L By Their Respective Dipeptidyl Glyoxal And Diazomethylketone Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-12-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: I0Y |
 |
Structural Basis For Irreversible Inhibition Of Human Cathepsin L By A Diazomethylketone Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-12-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: I0X |
 |
Crystal Structure Of Carcinoscorpius Rotundicauda Serine Protease Inhibitor Domain 1
Organism: Carcinoscorpius rotundicauda
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-08 Classification: HYDROLASE INHIBITOR |

