Search Count: 48
 |
Organism: Xenopus laevis, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: GENE REGULATION Ligands: ZSL, ZN |
 |
Organism: Claviceps fusiformis
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Structure Of Chanoclavine Synthase From Claviceps Fusiformis In Complex With Prechanoclavine
Organism: Claviceps fusiformis
Method: ELECTRON MICROSCOPY Resolution:2.33 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: HEM, LH6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-12-11 Classification: LIGASE Ligands: COA, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-12-11 Classification: LIGASE Ligands: COA, PO4, 2OP, MG |
 |
Beneficial Flip Of Substrate Orientation Enable Determine Substrate Specificity For Zearalenone Lactone Hydrolase
Organism: Exophiala aquamarina cbs 119918
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-12-06 Classification: HYDROLASE |
 |
Crystal Structure Of A Decarboxylase From Trichosporon Moniliiforme In Complex With O-Nitrophenol
Organism: Cutaneotrichosporon moniliiforme
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: OPO, MG |
 |
Cryo-Em Structure Of The Psi-Lhci-Lhcp Supercomplex From Ostreococcus Tauri
Organism: Ostreococcus tauri
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: PHOTOSYNTHESIS Ligands: CHL, CLA, IWJ, XAT, LHG, Q6L, BCR, LMG, SQD, CL0, PQN, SF4, DGD, NEX, KC2 |
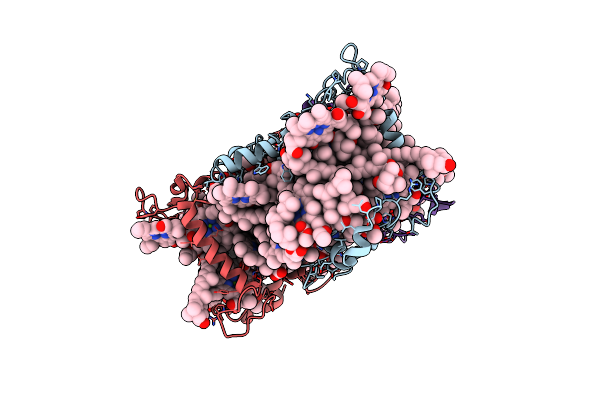 |
Organism: Ostreococcus tauri
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: ELECTRON TRANSPORT Ligands: CLA, CHL, KC2, Q6L, NEX, IWJ |
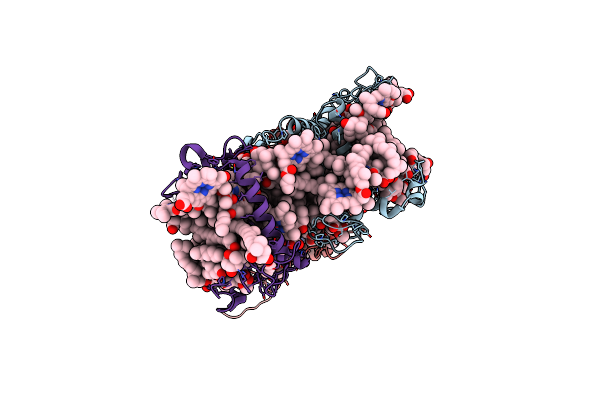 |
Cryo-Em Structure Of The Prasinophyte-Specific Light-Harvesting Complex (Lhcp)From Ostreococcus Tauri
Organism: Ostreococcus tauri
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: ELECTRON TRANSPORT Ligands: CLA, CHL, KC2, Q6L, NEX, IWJ |
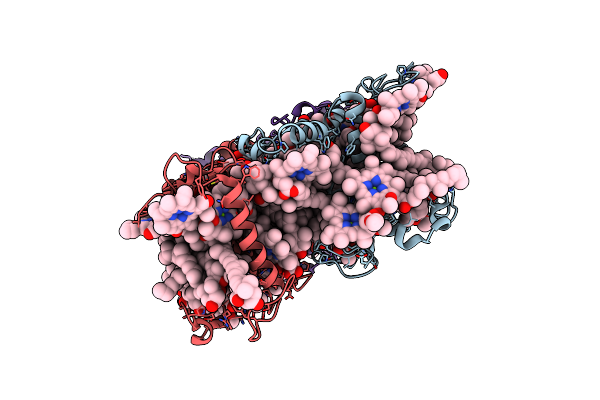 |
Cryo-Em Structure Of The Prasinophyte-Specific Light-Harvesting Complex (Lhcp)From Ostreococcus Tauri
Organism: Ostreococcus tauri
Method: ELECTRON MICROSCOPY Release Date: 2023-04-26 Classification: ELECTRON TRANSPORT Ligands: CLA, CHL, KC2, Q6L, IWJ, NEX |
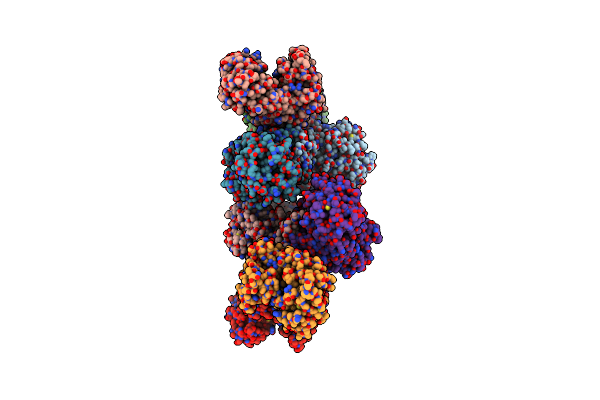 |
Complex Structure Of A Glucose 6-Phosphate Dehydrogenase From Zymomonas Mobilis
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE Ligands: NMN |
 |
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE |
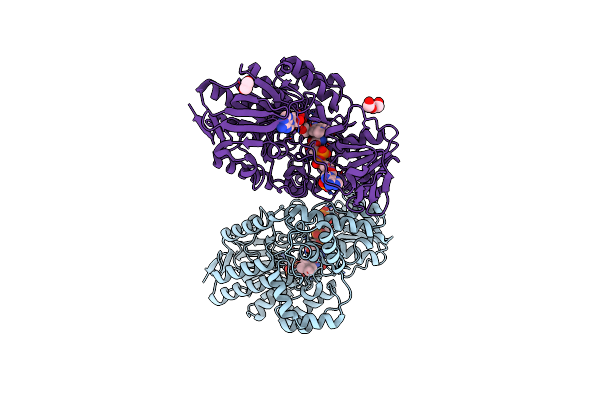 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P21221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, PEG, EDO, NA |
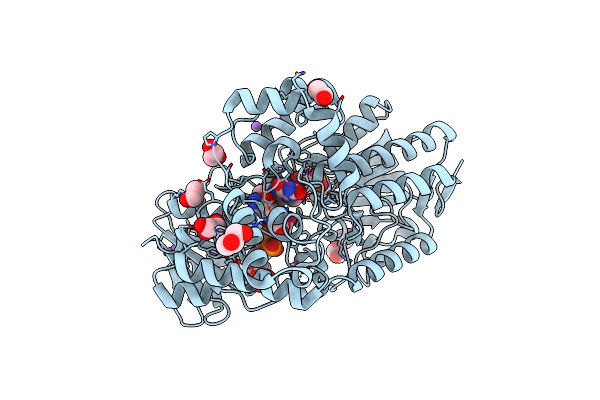 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of C2221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, EDO, GOL, NA |
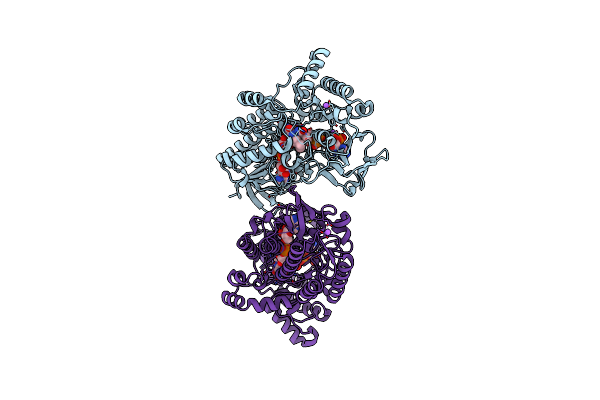 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P1211
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, NA |
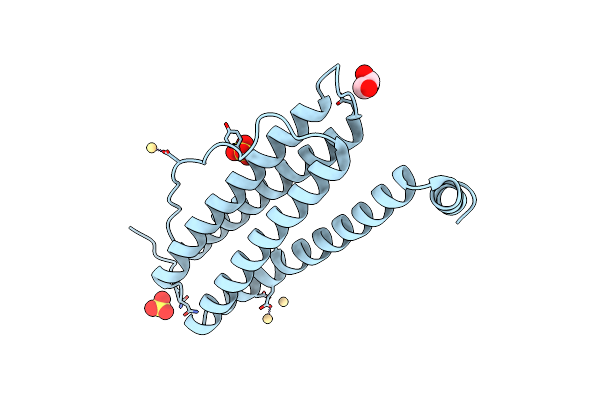 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-08 Classification: METAL BINDING PROTEIN Ligands: GOL, CD, SO4 |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-08 Classification: METAL BINDING PROTEIN Ligands: GOL, CD, AU, SO4 |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-08 Classification: METAL BINDING PROTEIN Ligands: EDO, AU, CD, SO4 |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-08 Classification: METAL BINDING PROTEIN Ligands: CD, SO4, AU |

