Search Count: 8
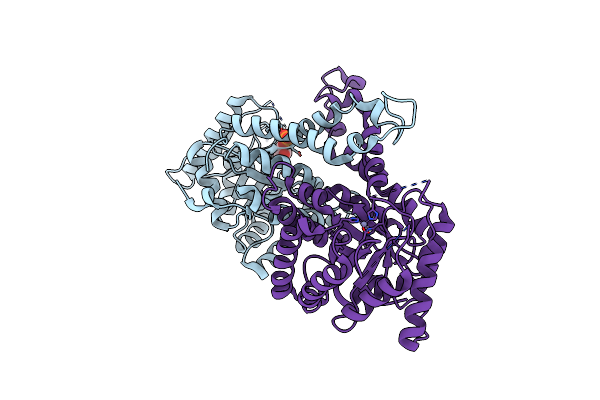 |
Structure Of Candida Albicans Fructose-1,6-Bisphosphate Aldolase Complexed With G3P
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-02-23 Classification: LYASE Ligands: ZN, G3H |
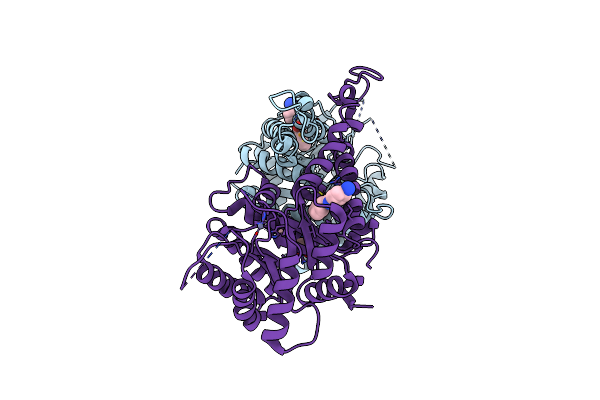 |
Structure Of Candida Albicans Fructose-1,6-Bisphosphate Aldolase Mutation C157S With Cn39
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-02-23 Classification: LYASE Ligands: ZN, EDO, 6Y3 |
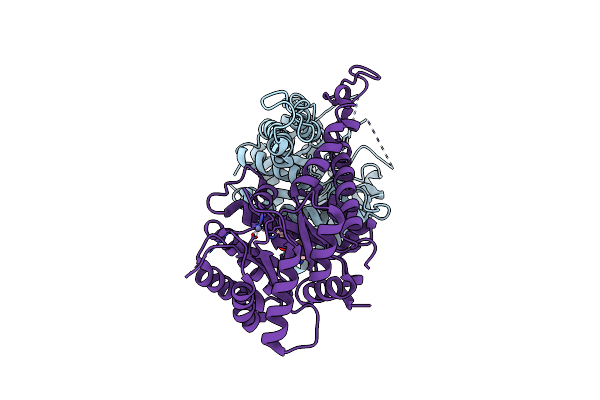 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-12-30 Classification: LYASE Ligands: ZN, EDO |
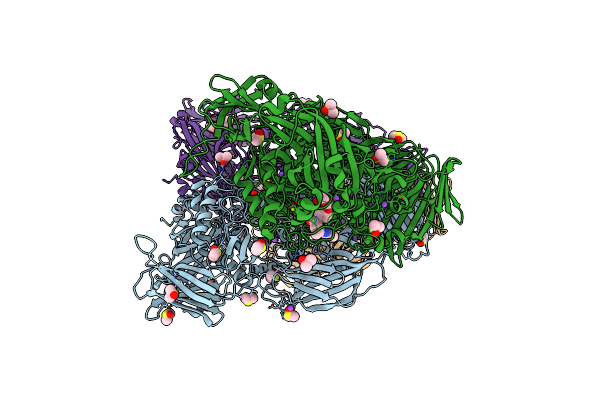 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: DVL, NA, DMS, MG, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-10-04 Classification: LIPID BINDING PROTEIN Ligands: P59, P69 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-10-04 Classification: LIPID BINDING PROTEIN Ligands: P59 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-10-04 Classification: LIPID BINDING PROTEIN Ligands: JAY |
 |
Crystal Structure Of The Mitotic Kinesin Eg5 In Complex With Mg-Adp And N-(3-Aminopropyl)-N-((3-Benzyl-5-Chloro-4-Oxo-3,4-Dihydropyrrolo[2,1-F][1,2,4]Triazin-2-Yl)(Cyclopropyl)Methyl)-4-Methylbenzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-06-27 Classification: CELL CYCLE Ligands: MG, 2AZ, ADP |

