Search Count: 36
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-06-19 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-06-19 Classification: FLAVOPROTEIN Ligands: FAD, CAA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-19 Classification: FLAVOPROTEIN Ligands: FAD, DAO |
 |
Structure Of Schistosoma Japonicum Glutathione S-Transferase Bound With The Ligand Complex Of 16
Organism: Schistosoma japonicum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-08-23 Classification: TRANSFERASE Ligands: 0IH, EOH |
 |
Organism: Helicobacter pylori (strain p12)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-11-09 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Helicobacter pylori (strain p12)
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2022-11-09 Classification: TRANSPORT PROTEIN Ligands: LW8 |
 |
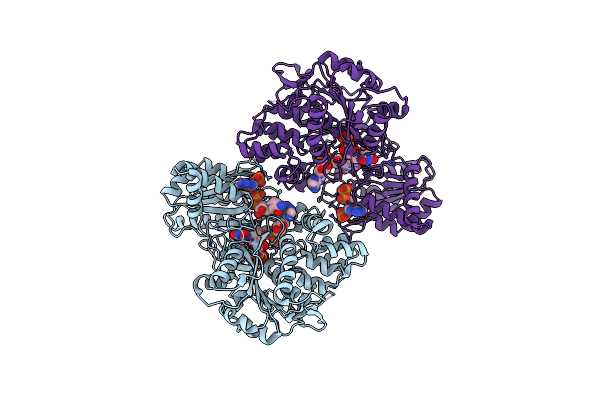
Structure Of The Cwld Amidase From Clostridioides Difficile In Complex With The Gers Lipoprotein
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: ZN, EDO |
 |
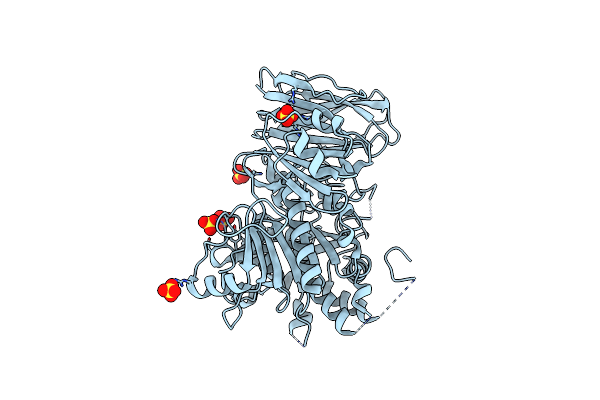
Crystal Structure Of A Tandem Deletion Mutant Of Rat Nadph-Cytochrome P450 Reductase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2021-01-13 Classification: FLAVOPROTEIN, OXIDOREDUCTASE Ligands: FMN, FAD, NAP |
 |
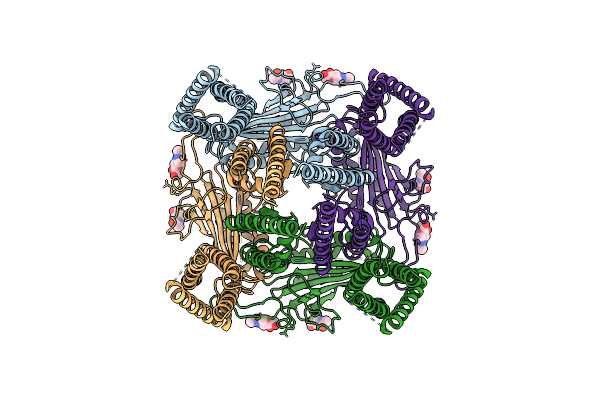
Organism: Peptoclostridium difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-06-26 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
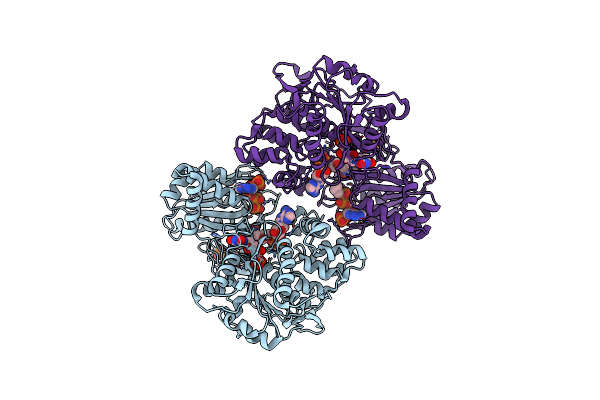
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: FMN, FAD, NAP |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2018-03-28 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FMN, FAD, NAP, PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FMN, FAD, NAP, PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FMN, FAD, NAP, PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FMN, FAD, NAP, PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FMN, FAD, 2AM, PO4 |
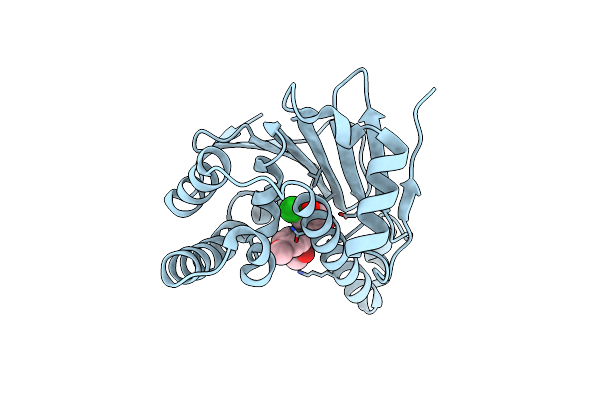 |
Crystal Structure Of The Human Hsp90-Alpha N-Domain Bound To The Hsp90 Inhibitor Fj2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-07-30 Classification: CHAPERONE Ligands: FJ2 |
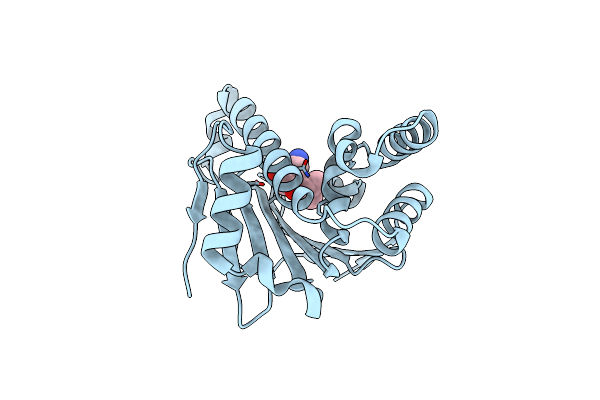 |
Crystal Structure Of The Human Hsp90-Alpha N-Domain Bound To The Hsp90 Inhibitor Fj3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-07-30 Classification: CHAPERONE Ligands: FJ3 |
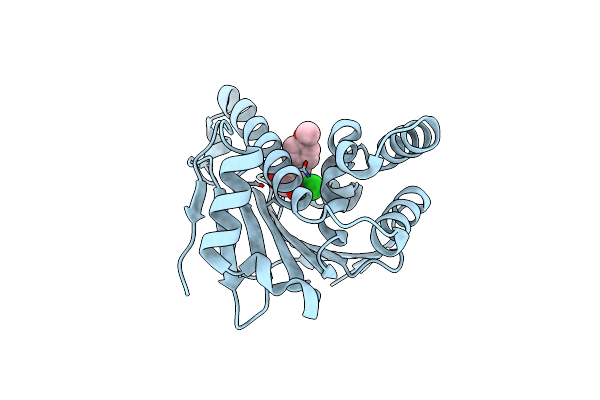 |
Crystal Structure Of The Human Hsp90-Alpha N-Domain Bound To The Hsp90 Inhibitor Fj4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-07-30 Classification: CHAPERONE Ligands: FJ4 |
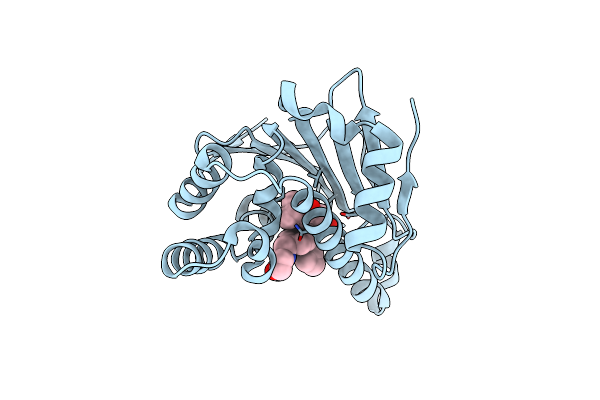 |
Crystal Structure Of The Human Hsp90-Alpha N-Domain Bound To The Hsp90 Inhibitor Fj5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-07-30 Classification: CHAPERONE Ligands: FJ5 |

