Search Count: 16
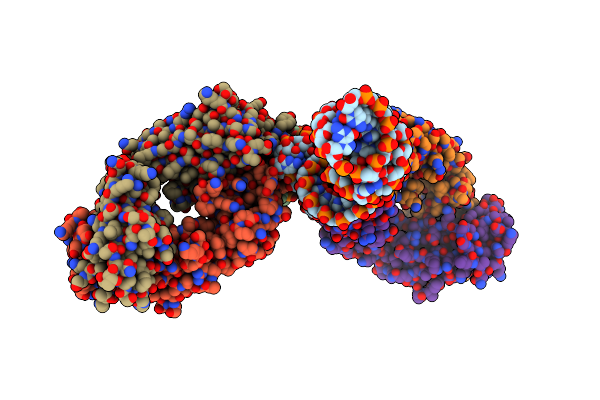 |
Crystal Structure Of A Self-Alkylating Ribozyme - Alkylated Form With Biotinylated Epoxide Substrate
Organism: Homo sapiens, Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-01-19 Classification: RNA/IMMUNE SYSTEM Ligands: V4J |
 |
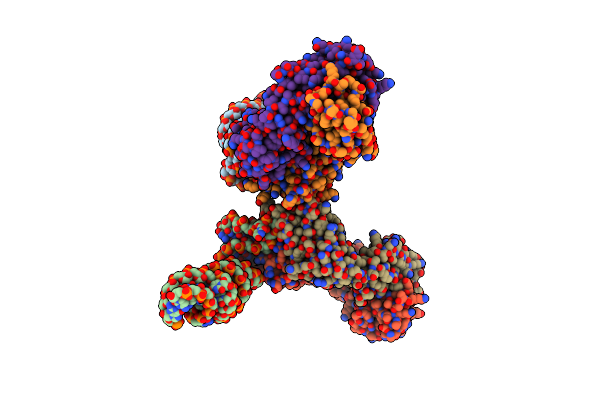
Crystal Structure Of A Self-Alkylating Ribozyme - Alkylated Form Without Biotin Moiety
Organism: Homo sapiens, Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-01-19 Classification: RNA/IMMUNE SYSTEM Ligands: 97C |
 |
Crystal Structure Of A Self-Alkylating Ribozyme - Short Time Incubation With The Epoxide Substrate
Organism: Homo sapiens, Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-01-19 Classification: RNA/IMMUNE SYSTEM |
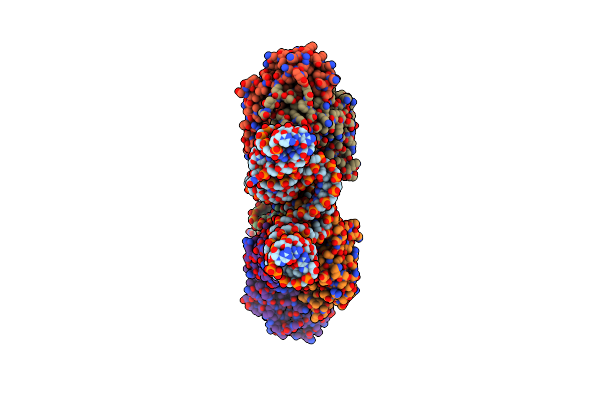
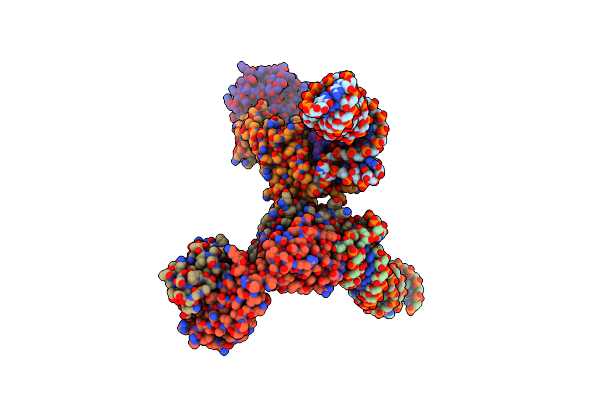 |
Organism: Homo sapiens, Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-01-19 Classification: RNA/IMMUNE SYSTEM |
 |
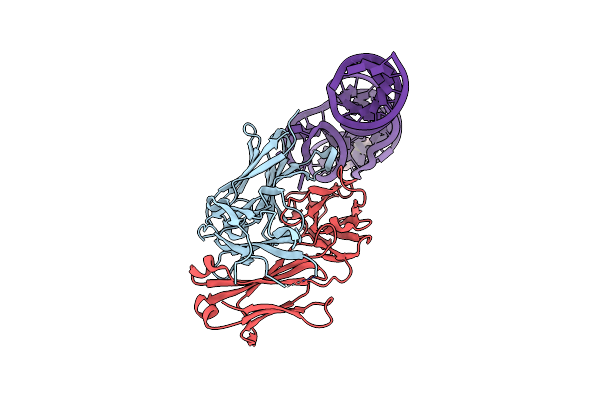
Crystal Structure Of Hepatitis A Virus Ires Domain V In Complex With Fab Havx
Organism: Homo sapiens, Hepatovirus a
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-08-14 Classification: RNA/Immune System |
 |
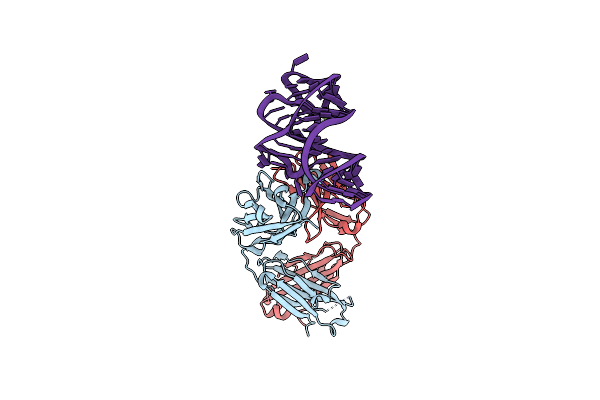
Structural Basis For Promiscuous Binding And Activation Of Fluorogenic Dyes By Dir2S Rna Aptamer
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2018-11-14 Classification: RNA/IMMUNE SYSTEM Ligands: G4A |
 |
Structural Basis For Promiscuous Binding And Activation Of Fluorogenic Dyes By Dir2S Rna Aptamer
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-11-14 Classification: RNA/Immune System Ligands: MG |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2017-12-27 Classification: immune system/rna Ligands: MG |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2017-12-27 Classification: immune system/rna Ligands: MG |
 |
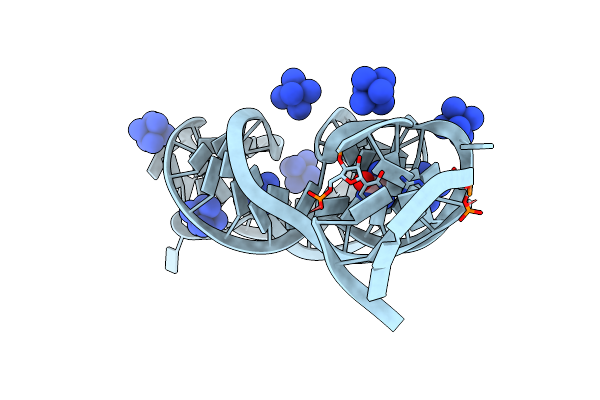
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2015-08-12 Classification: RNA Ligands: NCO, HPA |
 |
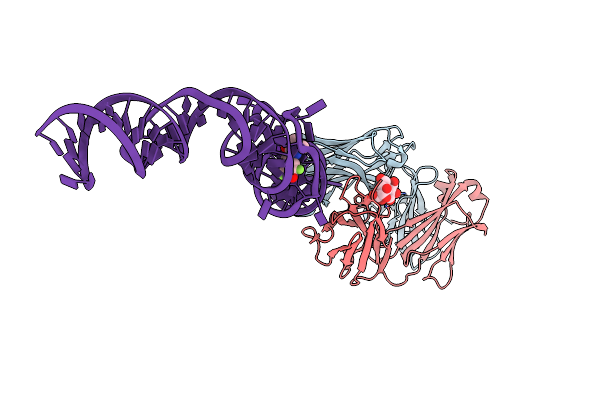
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2015-08-12 Classification: RNA Ligands: NCO, HPA |
 |
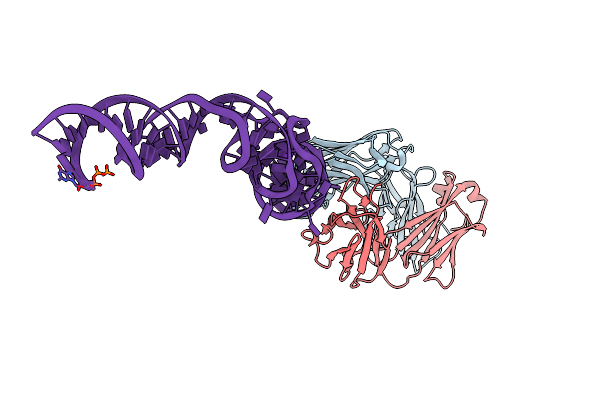
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-06-18 Classification: immune system/rna Ligands: K, MG, 1TU |
 |
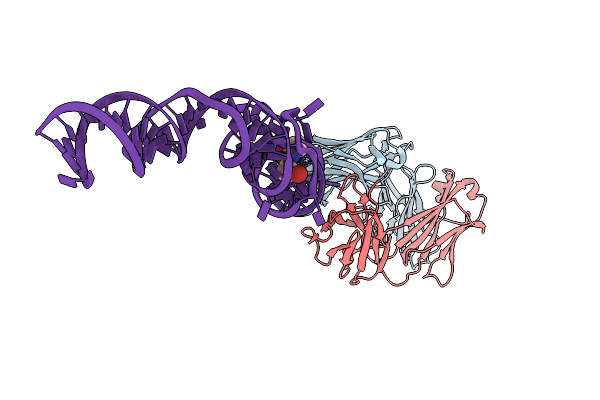
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-06-18 Classification: immune system/rna Ligands: K, MG |
 |
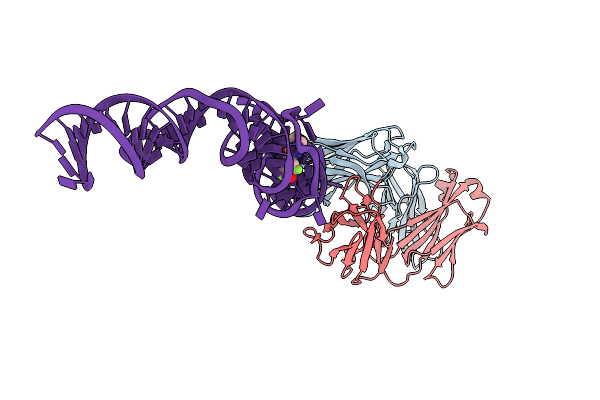
Crystal Structure Of An Rna Aptamer Bound To Bromo-Ligand Analog In Complex With Fab
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-06-18 Classification: RNA Ligands: MG, K, 2ZZ |
 |
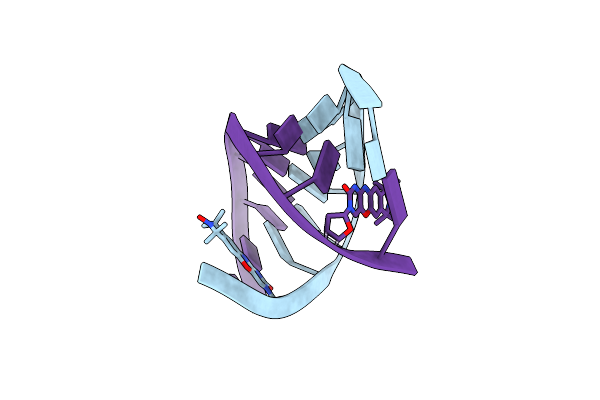
Crystal Structure Of An Rna Aptamer Bound To Trifluoroethyl-Ligand Analog In Complex With Fab
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2014-06-18 Classification: RNA/immune system Ligands: K, 2ZY |
 |
Crystal Structure Of A Dna Containing The Rigid Nitroxide Spin-Labeled Nucleotide C-Spin
|

