Search Count: 136
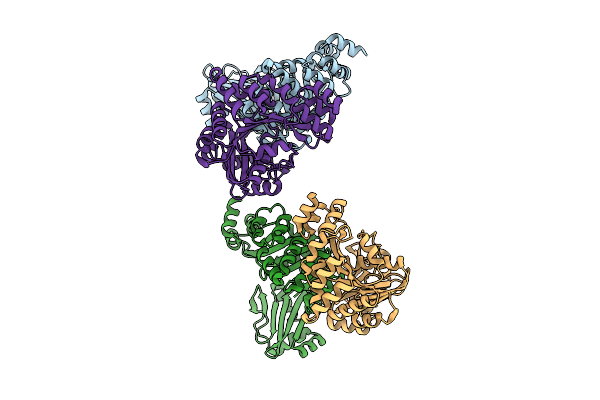 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: MOTOR PROTEIN |
 |
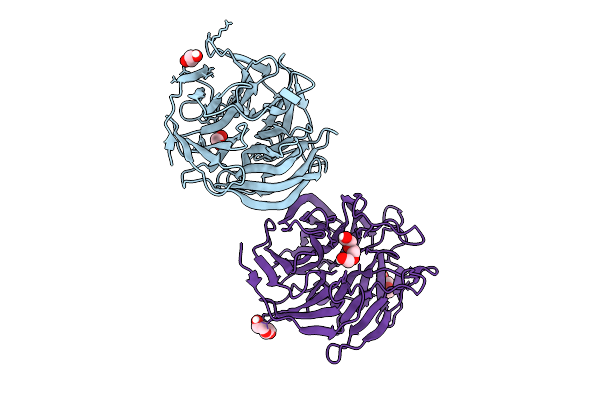
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: APOPTOSIS |
 |
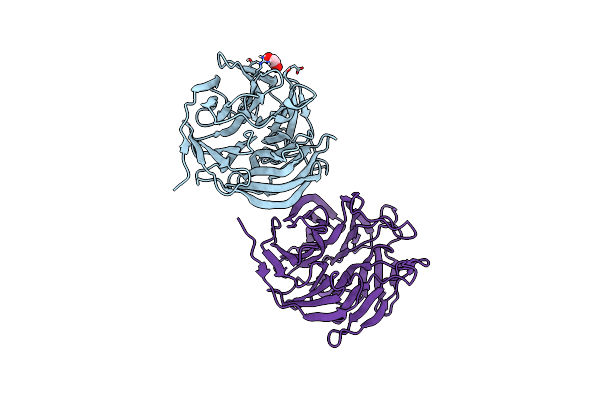
Crystal Structure Of Complement1.4 (Cent1.4), An Engineered Photoenzyme For Selective [2+2]-Cycloadditions
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, PEG, PGE |
 |
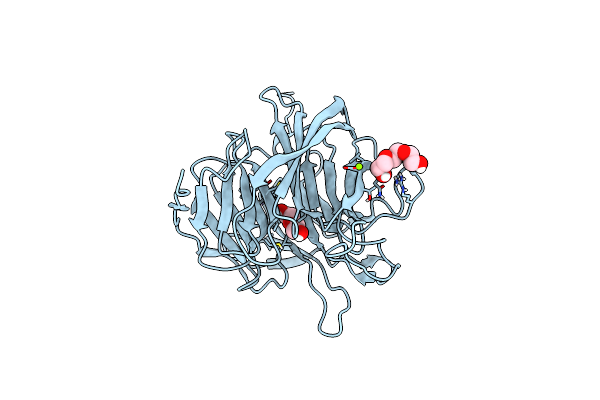
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, EDO |
 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PGE, MG |
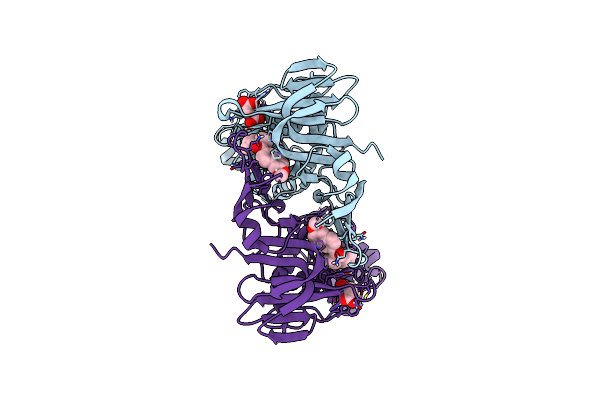 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-04-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A0T, SIN |
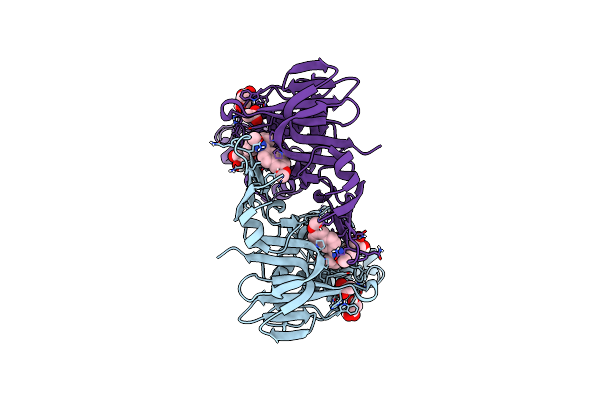 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-04-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A0S, ACY, GOL |
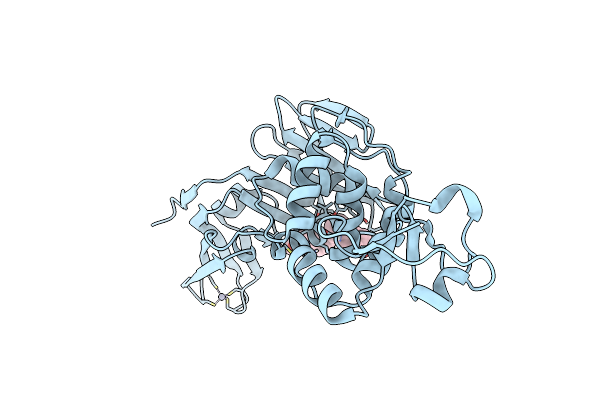 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-04-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A0U, ZN, CL |
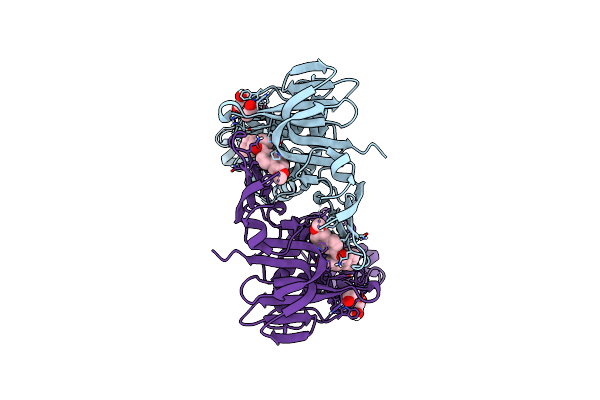 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A0V, ACY, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Bhmehis1.8, An Engineered Enzyme For The Morita-Baylis-Hillman Reaction
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PGE, EDO |
 |
Crystal Structure Of Bhmehis1.0, An Engineered Enzyme For The Morita-Baylis-Hillman Reaction
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-02-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, ACT, MG, SO4 |
 |
Organism: Legionella pneumophila 130b
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-12-27 Classification: CELL ADHESION |
 |
Structure Of Legionella Pneumophila Lcl C-Terminal Domain Bound To Sulphate
Organism: Legionella pneumophila 130b
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-12-27 Classification: CELL ADHESION Ligands: SO4 |
 |
Crystal Structure Of A Computationally Designed Heme Binding Protein, Dnhem1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-05 Classification: BIOSYNTHETIC PROTEIN Ligands: IMD, HEM, MPD, PO4, PEG, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN Ligands: PO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Evolved Photoenzyme Ent1.3 (Truncated) With Bound Product
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-28 Classification: BIOSYNTHETIC PROTEIN Ligands: JRP, EDO |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: NAG, ACT, GOL |

