Search Count: 239
 |
Crystal Structure Of The Chymotrypsin-Cleaved Iron-Free C-Lobe Of Bovine Lactoferrin At 2.82 Angstrom Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2025-12-31 Classification: METAL BINDING PROTEIN Ligands: EDO, GOL, ACT, NAG, SO4 |
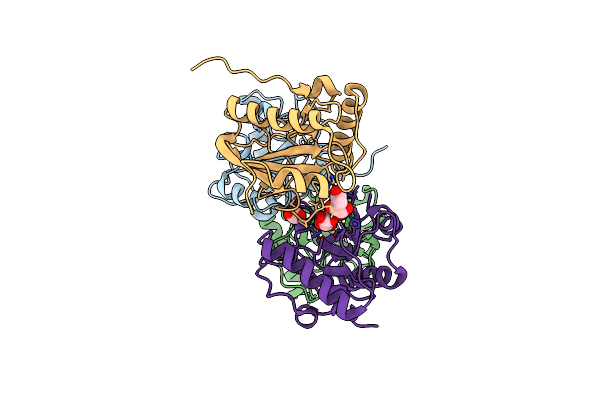 |
Crystal Structure Of The Complex Of Camel Peptidoglycan Recognition Protein, Pgrp-S With Malic Acid And Oxalic Acid At 2.3 A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2025-05-14 Classification: IMMUNE SYSTEM Ligands: MLT, OXD |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The 17-Mer Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.10 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: CIT |
 |
Crystal Structure Of The Complex Of Lactoperoxidase With Nitric Oxide At 1.72 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: NAG, IOD, SCN, PGE, NO2, NO, HEM, CA |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.20 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: PAE, GOL |
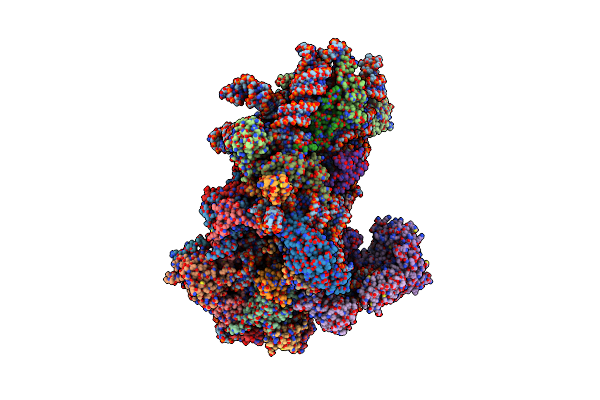 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
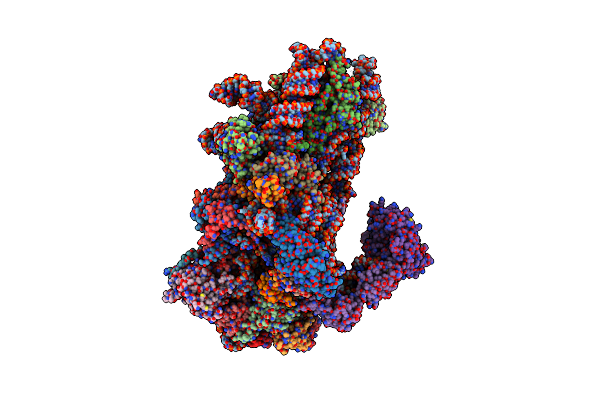
Amg 193, A Clinical Stage Mta-Cooperative Prmt5 Inhibitor, Drives Anti-Tumor Activity Preclinically And In Patients With Mtap-Deleted Cancers
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GOL, DMS, A1ATH, MTA |
 |
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
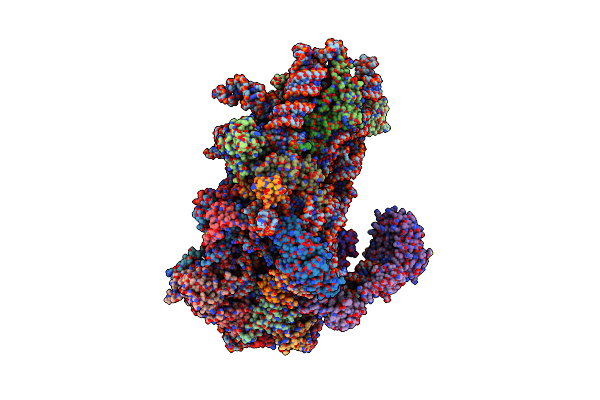
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif5)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
 |
Crystal Structure Of The Ternary Complex Of Lactoperoxidase With Nitric Oxide And Nitrite Ion At 1.95 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE Ligands: NAG, NO, NO2, NO3, SCN, IOD, HEM, EDO, CA, OSM |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.39 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.37 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The 17-Mer Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.60 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: CIT, GOL |
 |
Structure Of Phosphopantetheine Adenylyltransferase (Ppat) From Enterobacter Spp. With The Expression Tag Bound In The Substrate Binding Site Of A Neighbouring Molecule At 2.25 A Resolution.
Organism: Enterobacter sp. 638
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: PAE, GOL, EDO |

