Search Count: 158
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-B-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
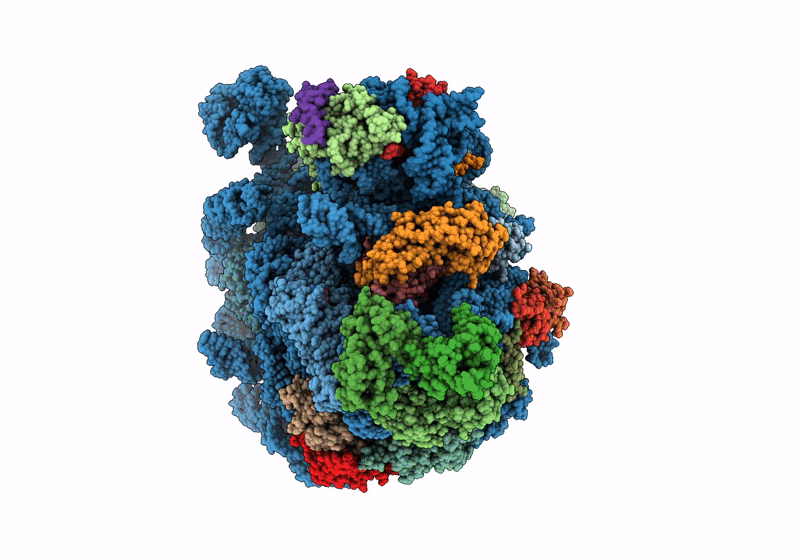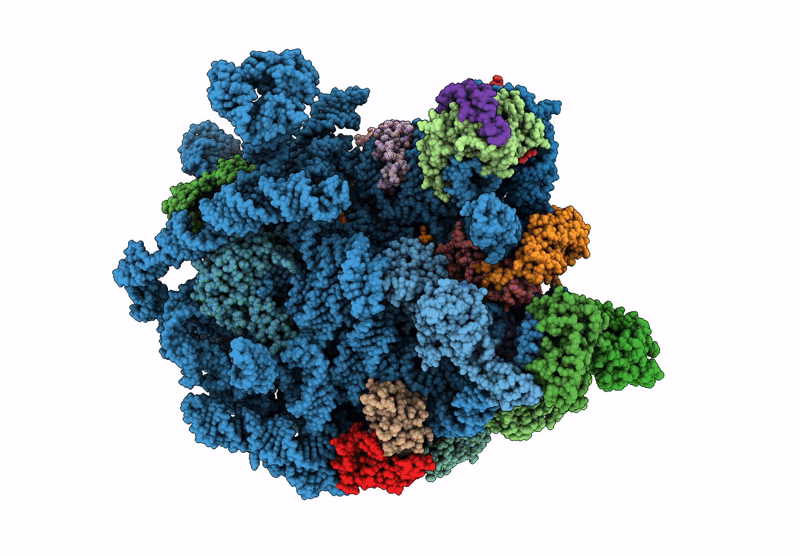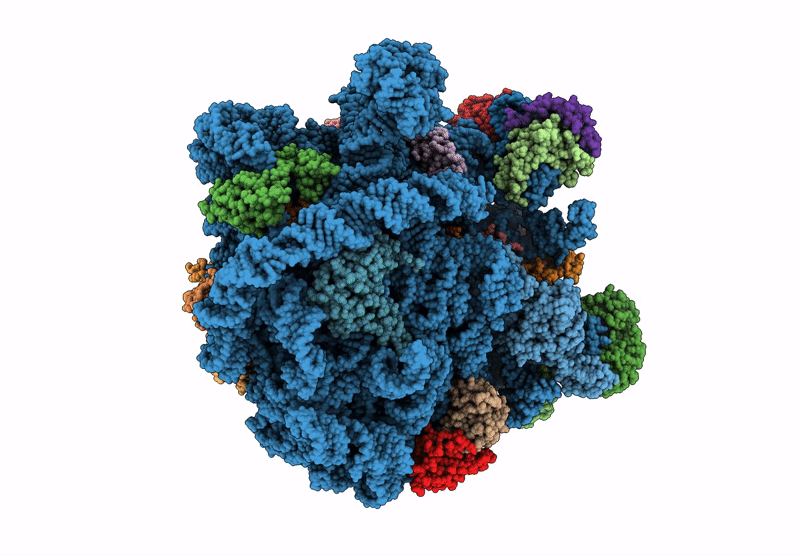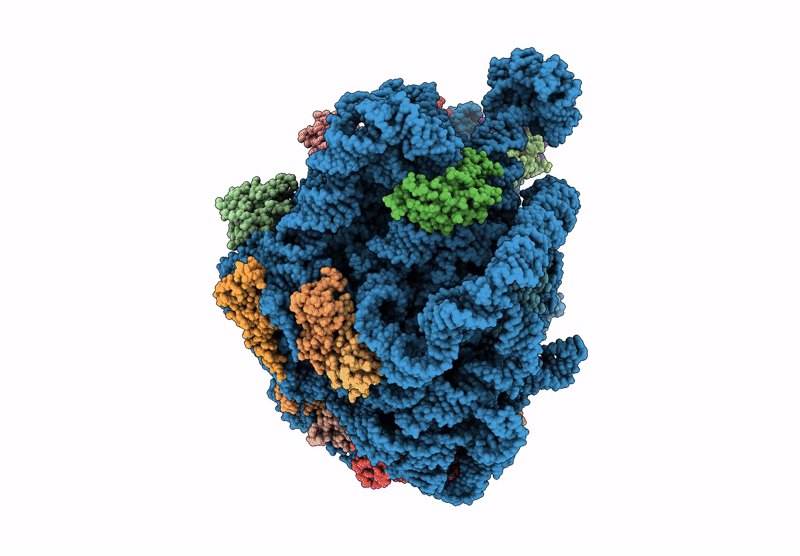
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-A
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-B
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-C
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Delnte-Hflx
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME |
 |
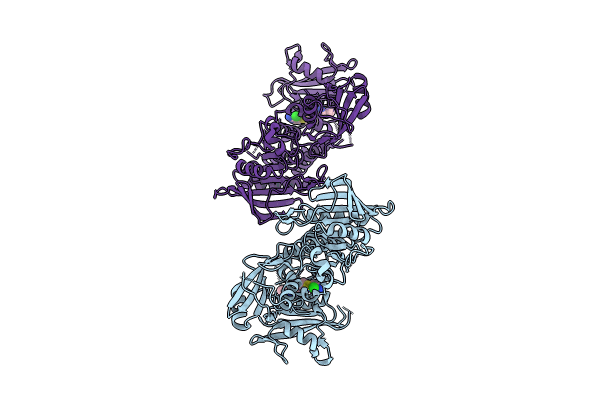
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-A-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Chloramphenicol:50S-Hflx-B-Clm
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, CLM |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Chloramphenicol:50S-Hflx-A-Clm
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, CLM |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE |
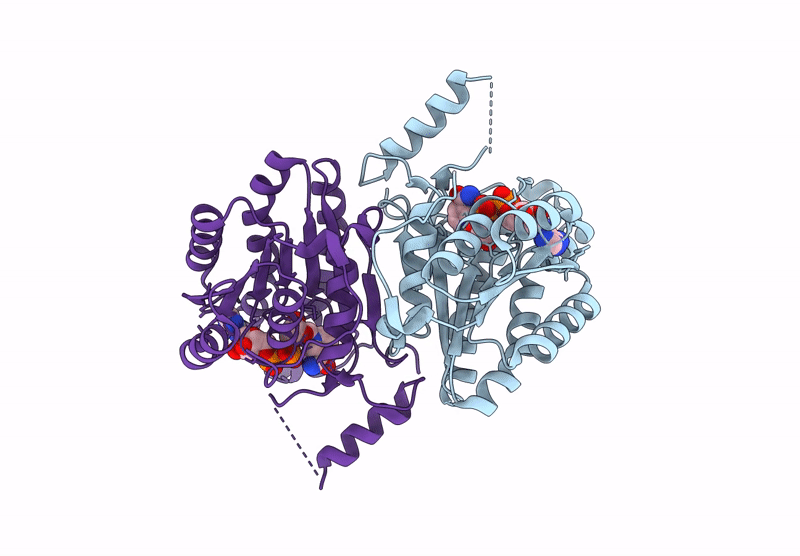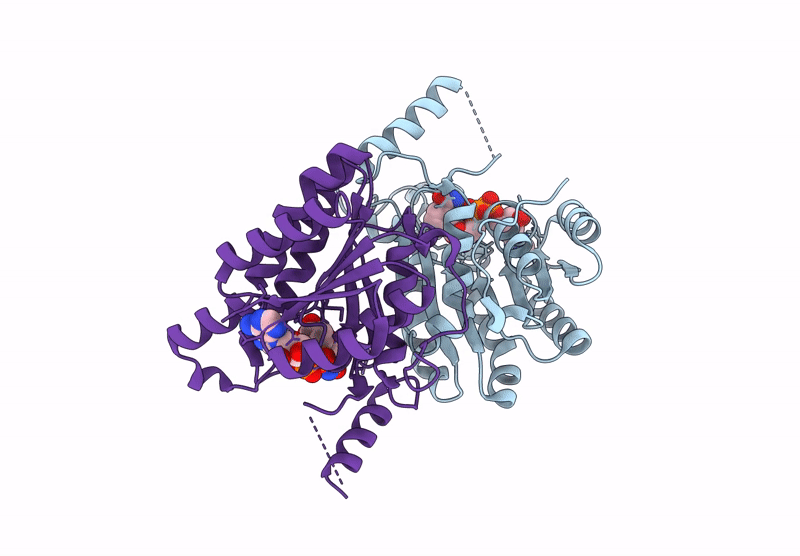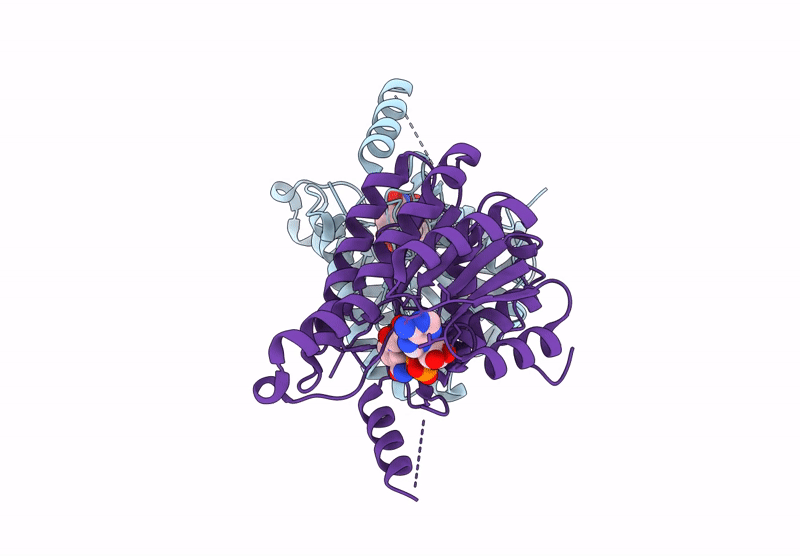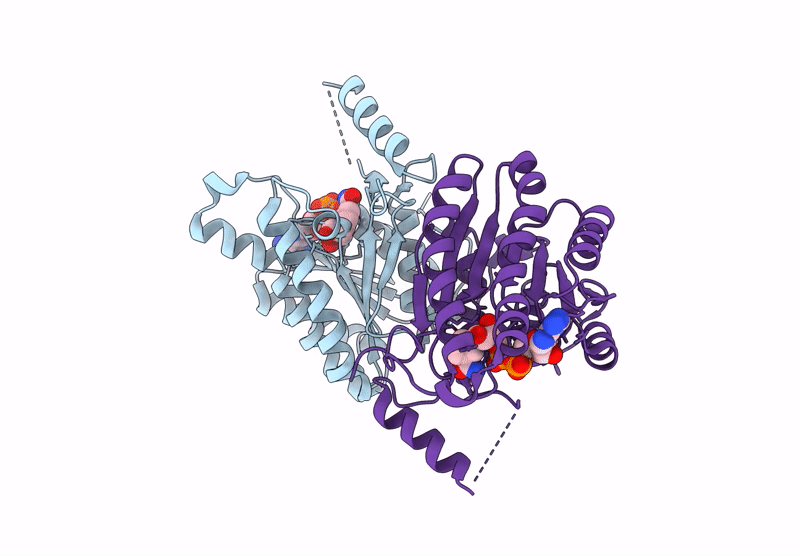 |
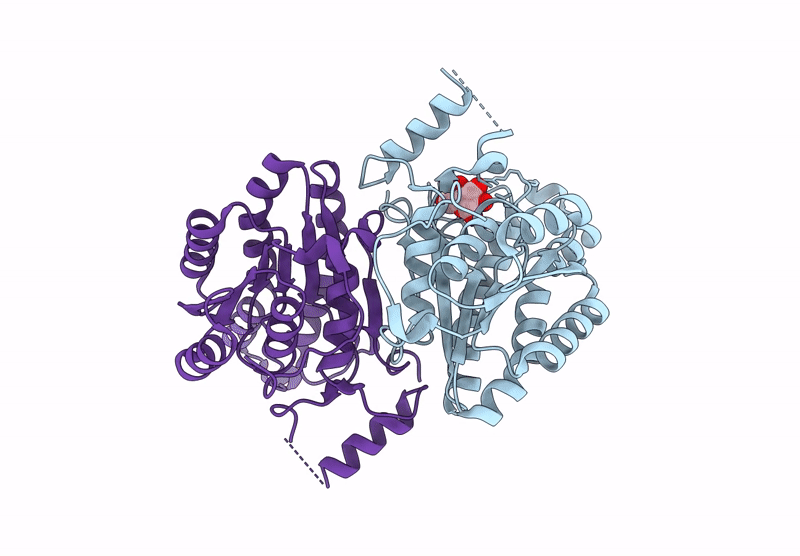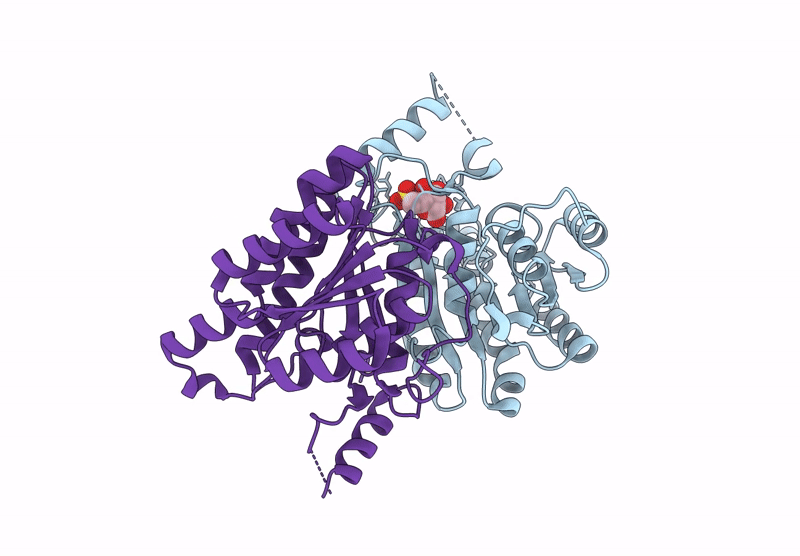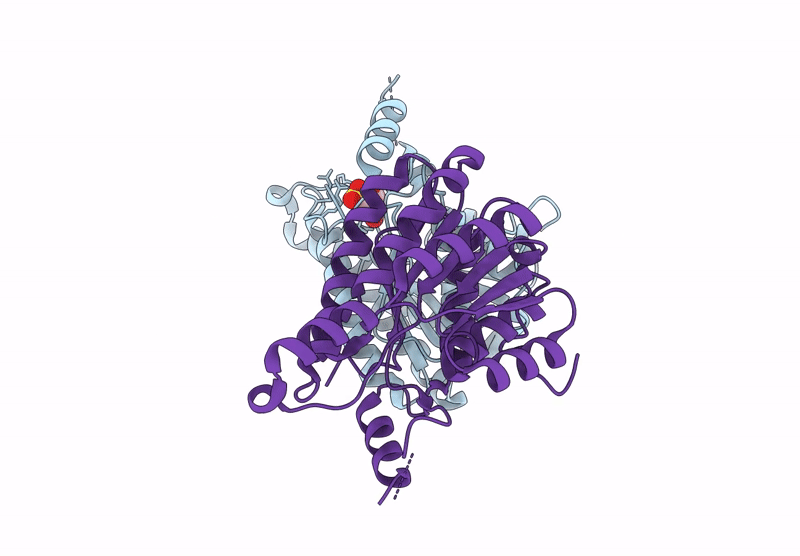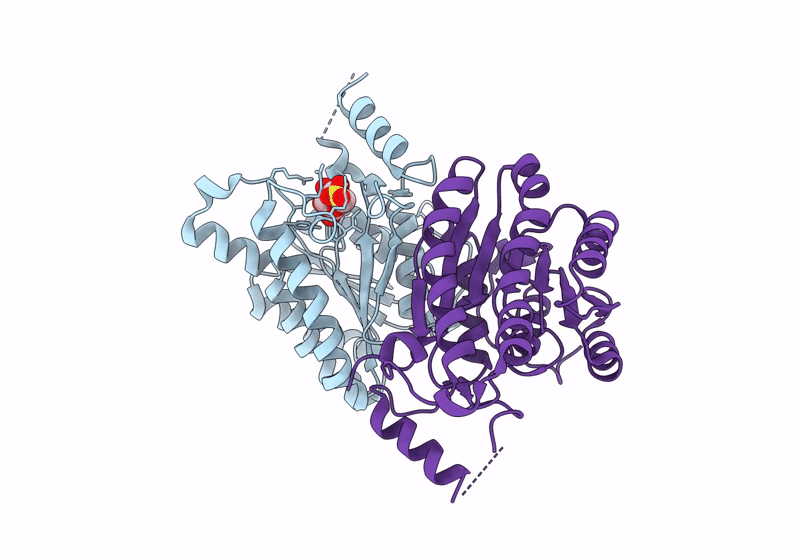
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Nad+ (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Sulfoquinovose Substrate (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: YZT |
 |
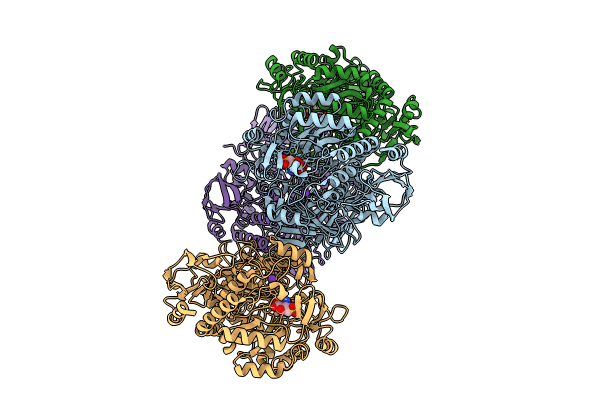
Crystal Structure Of Non-Receptor Protein Tyrosine Phosphatase Shp2 In Complex With Pf-07284892
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-05-29 Classification: HYDROLASE/INHIBITOR Ligands: A1AQ1 |
 |
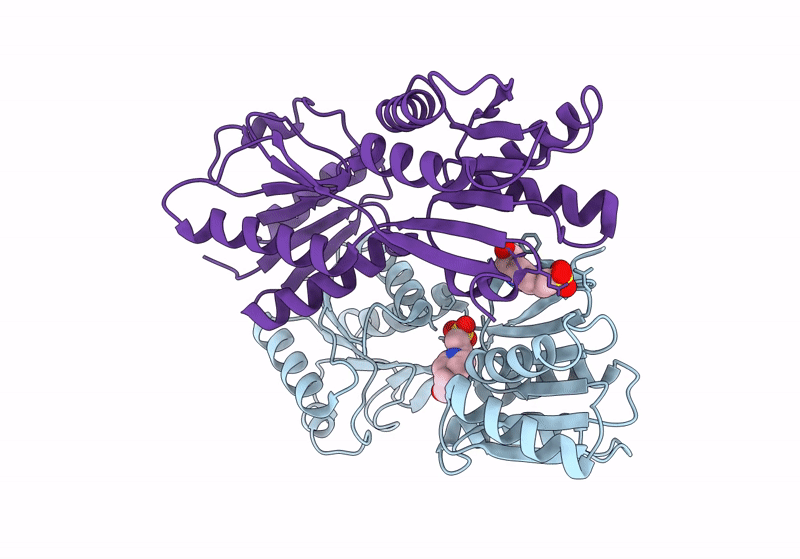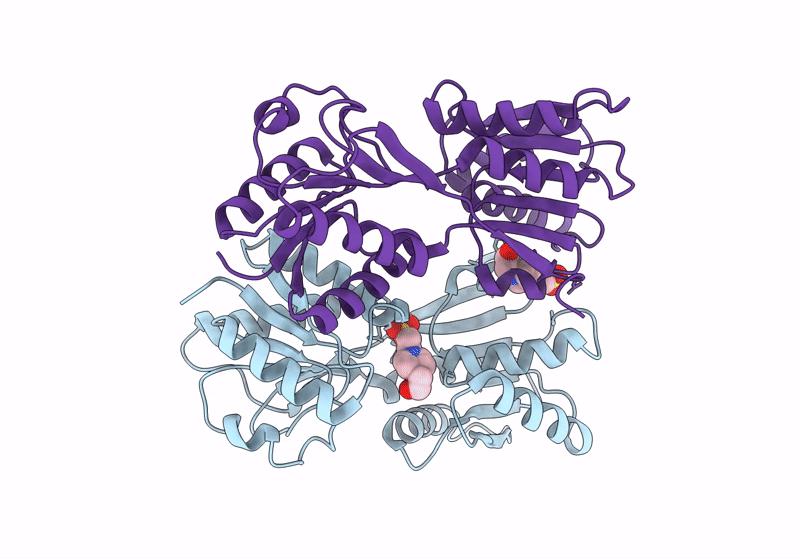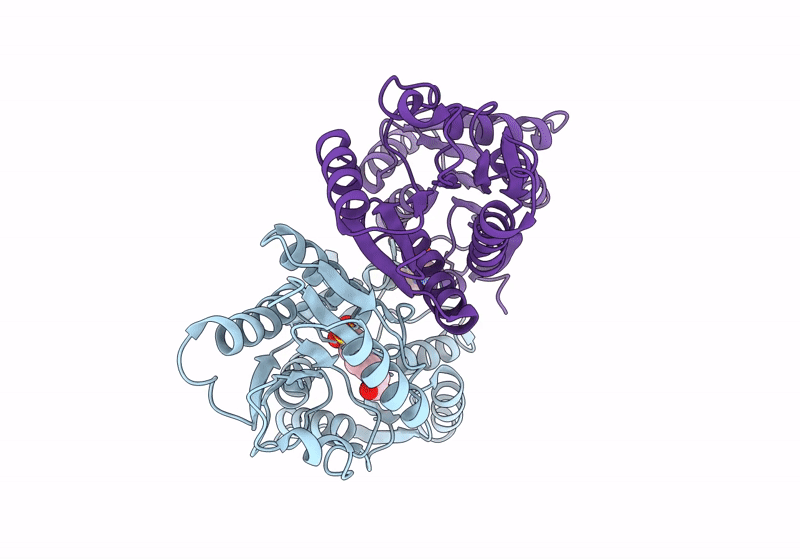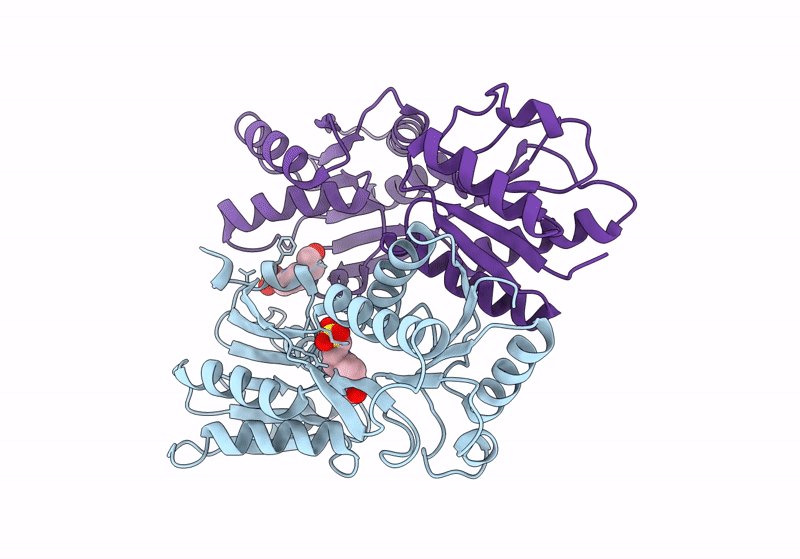
Crystal Structure Of Gh31 Family Sulfoquinovosidase Bmsqase In Covalent Complex With Sq-Aziridine (Sqz)
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: Y2W, K |
 |
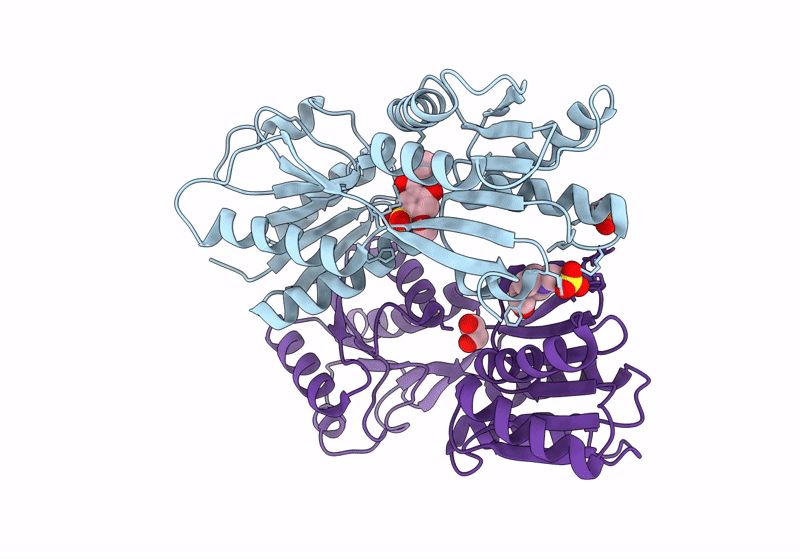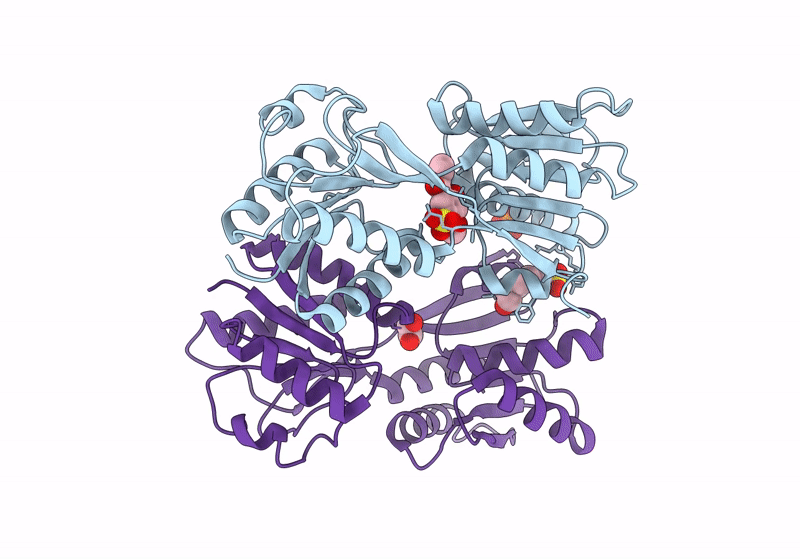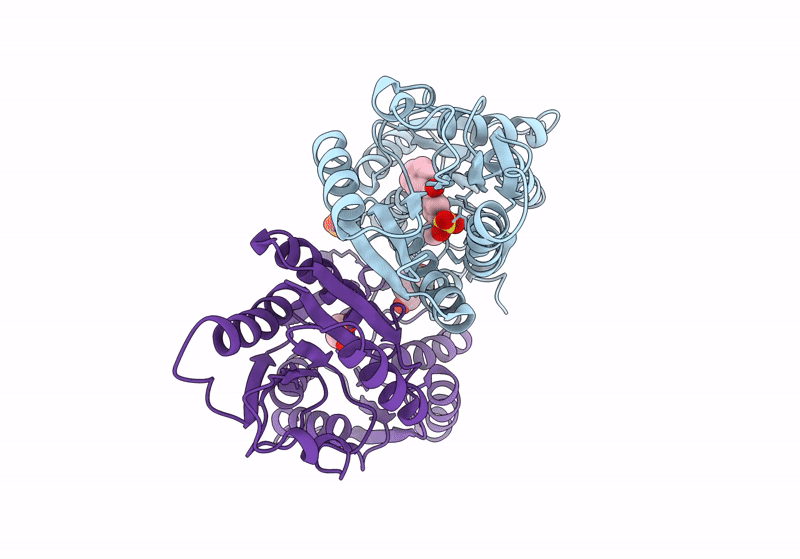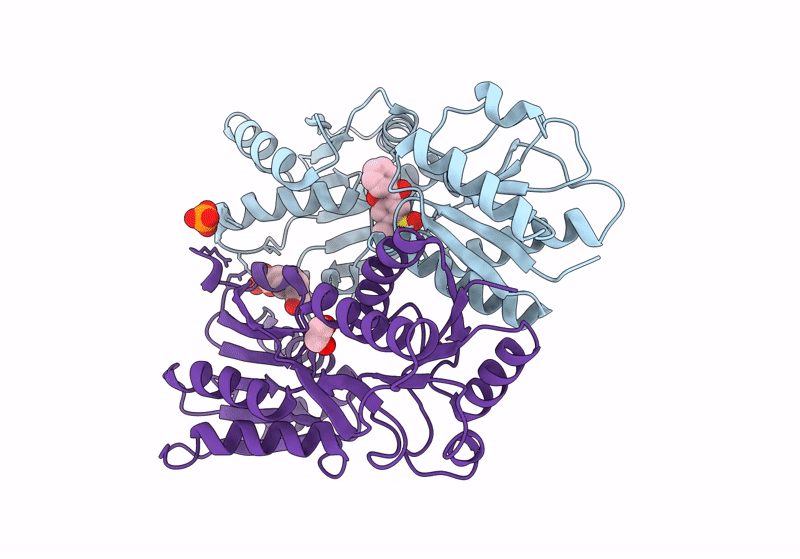
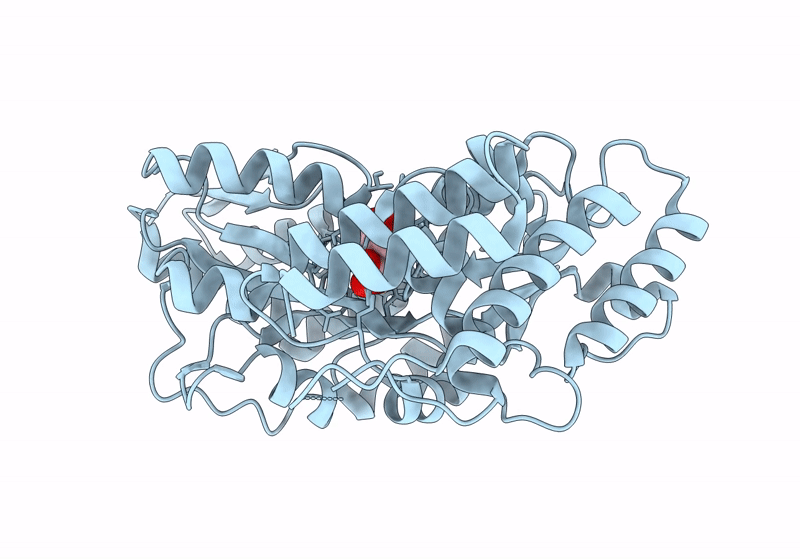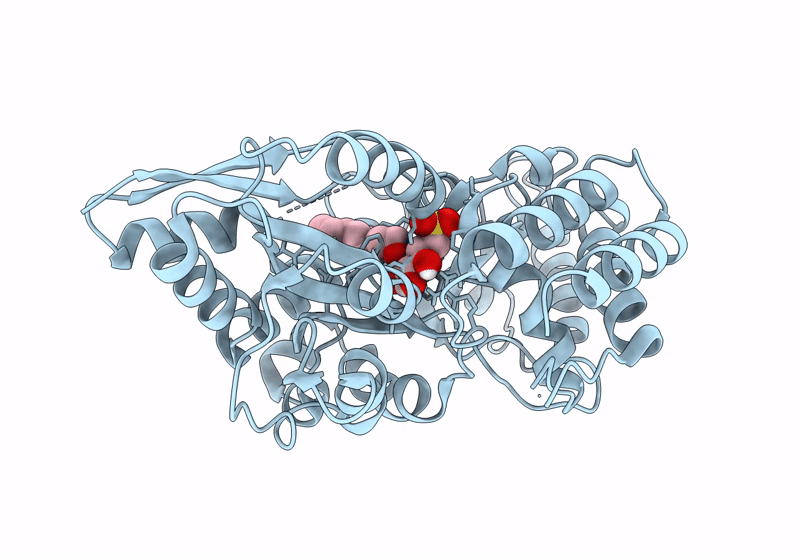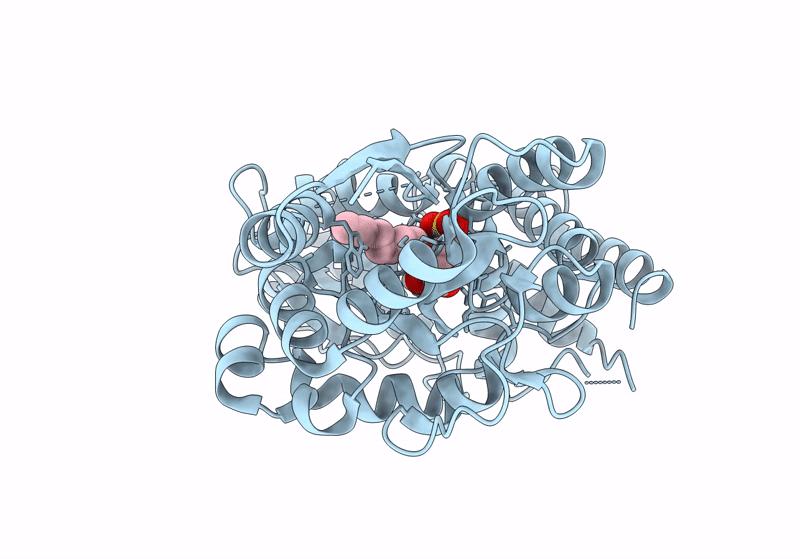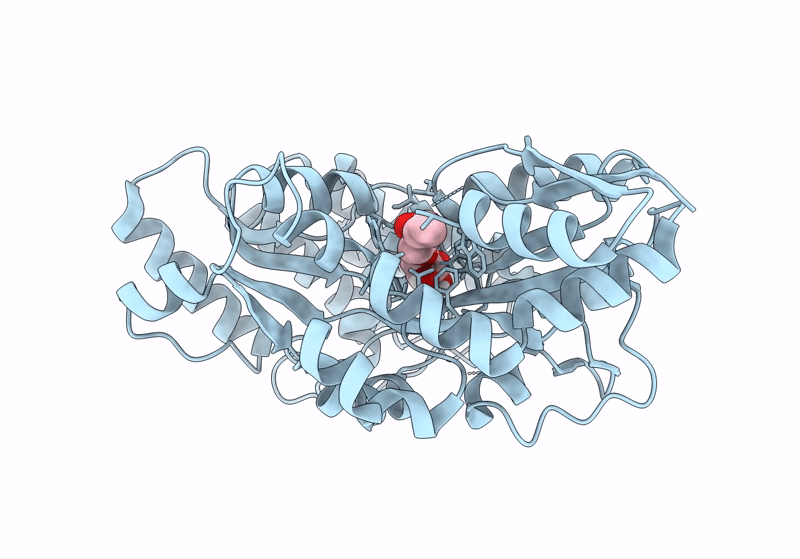
Crystal Structure Of Catabolite Repressor Acivator From E. Coli In Complex With Hepes
Organism: Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2024-04-24 Classification: GENE REGULATION Ligands: EPE |
 |
Crystal Structure Of Catabolite Repressor Acivator From E. Coli In Complex With Sulisobenzone
Organism: Escherichia coli 536
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: I4Y, PO4, EPE, EDO |
 |
Crystal Structure Of The Sulfoquinovosyl Binding Protein (Smof) From A. Tumefaciens Sulfo-Smo Pathway In Complex With Sqoctyl Ligand
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-04-17 Classification: SUGAR BINDING PROTEIN Ligands: A1H5E |

