Search Count: 185
 |
The Crystal Structure Of Covid-19 Main Protease In Complex With An Inhibitor Minocycline
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-02-12 Classification: ANTIVIRAL PROTEIN Ligands: MIY |
 |
Solution Structure Of Mu-Theraphotoxin Cg4A From Chinese Tarantula Chilobrachys Jingzhao
Organism: Chilobrachys guangxiensis
Method: SOLUTION NMR Release Date: 2023-11-08 Classification: TOXIN |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: PEG, TCH |
 |
Crystal Structure Of Vibrio Cholerae Dsba In Complex With Bile Salt Taurocholate
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-12-29 Classification: OXIDOREDUCTASE Ligands: GOL, TCH |
 |
Crystal Structure Of O-Acetyl L-Serine Sulfhydrylase From Haemophilus Influenzae In Complex With C-Terminal Peptide Of Ribosomal S4 Domain Protein From Lactobacillus Salivarius.
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd), Lactobacillus salivarius ucc118
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-12-09 Classification: TRANSFERASE |
 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product F1 (Methylpiperazinone 6)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O7P, CU, EDO |
 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product G6 (Pyrazole 9)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAV, CU |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-06-24 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: KFS, CU |
 |
Crystal Structure Of Ecdsba In A Complex With Purified Methylpiperazinone 6
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: O7P, PGE |
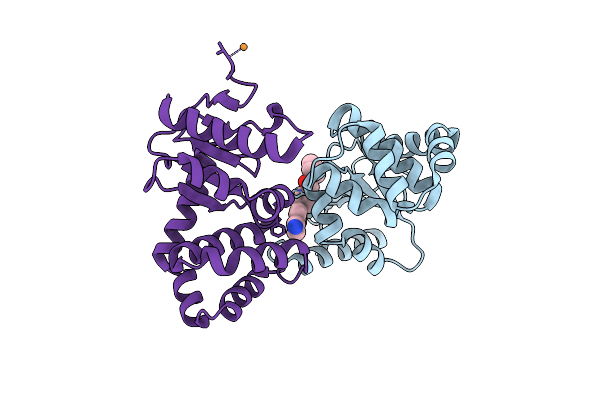 |
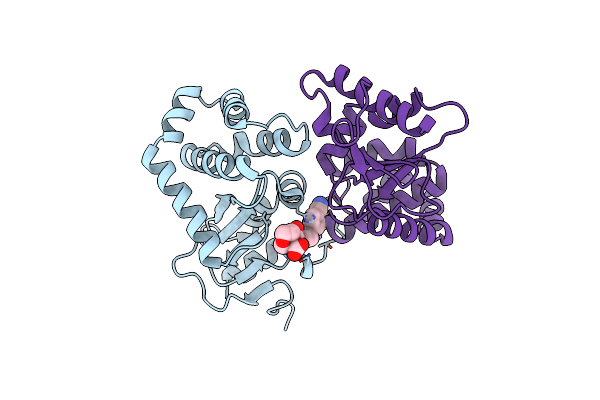
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O6Y, CU |
 |
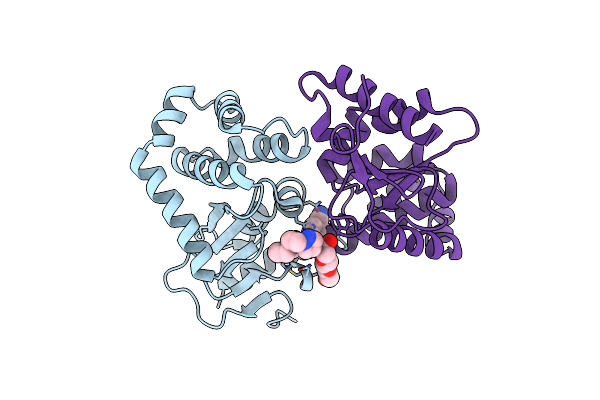
Crystal Structure Of Ecdsba In A Complex With Purified Morpholine Carboxylic Acid 7
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAJ |
 |
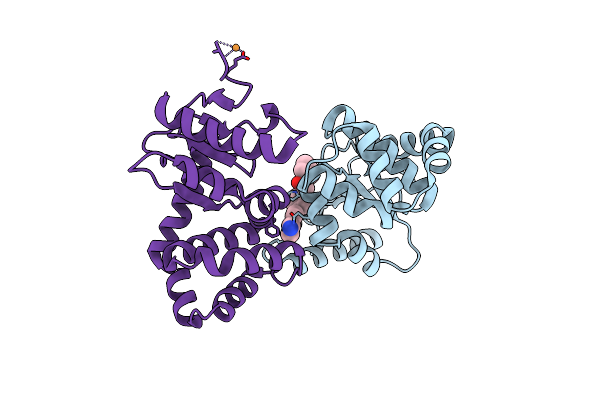
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAV, PGE |
 |
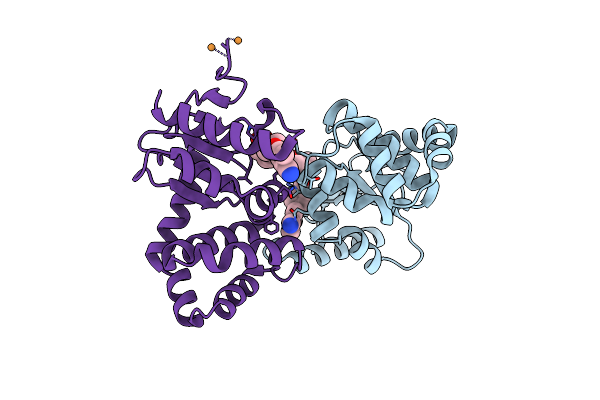
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product H5 (Morpholine 8)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O6Y, CU |
 |
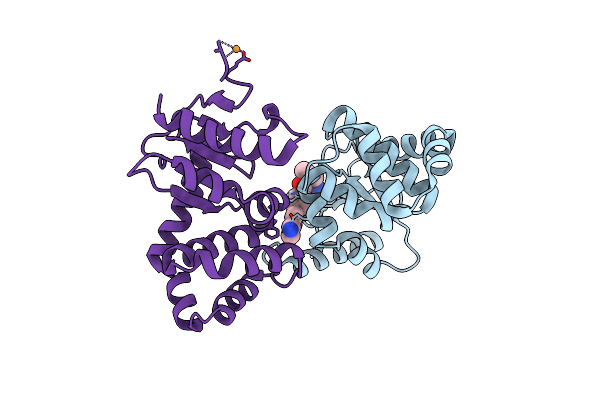
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product A5 (Morpholine Carboxylic Acid 7)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OMJ, CU, OAJ |
 |
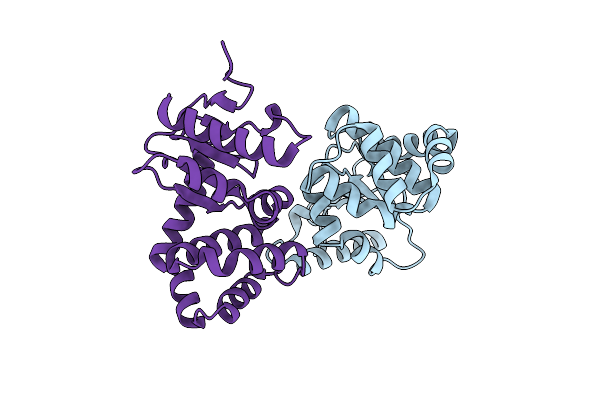
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: ONY, CU |
 |
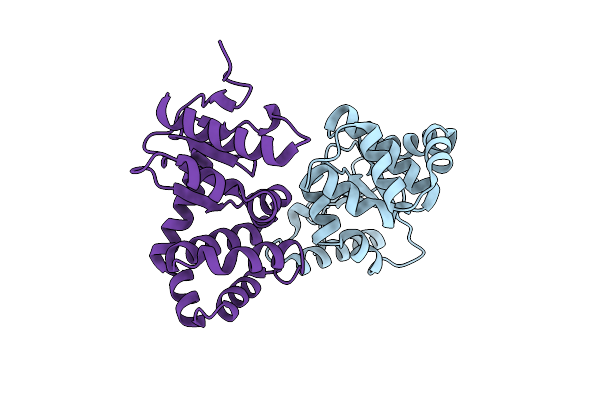
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A1_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A10_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A11_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A12_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A2_3)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |

