Search Count: 47
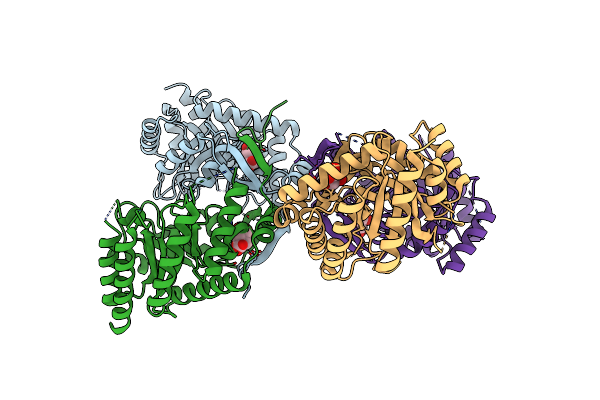 |
Crystal Structure Of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase (Dahp Synthase) From Providencia Alcalifaciens Complexed With Quinic Acid
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: QIC |
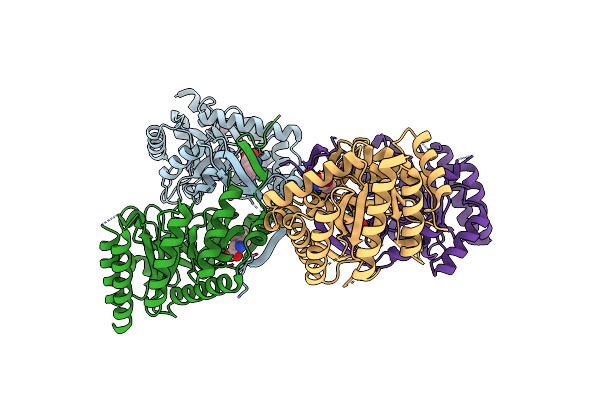 |
Crystal Structure Of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase (Dahp Synthase) From Providencia Alcalifaciens Complexed With Phe
Organism: Providencia alcalifaciens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: PEG, PHE |
 |
Crystal Structure Of Active Kras-G12C (Gmppnp-Bound) In Complex With Bbo-8520
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GNP, MG, Y8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2024-12-18 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, Y8N |
 |
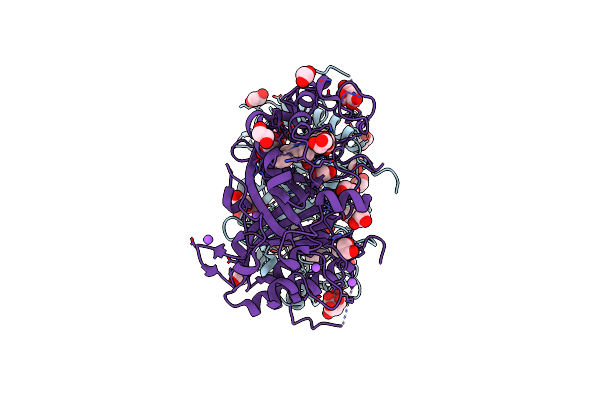
Crystal Structure Of Haloacid Dehalogenase-Like Hydrolase Family Enzyme From Staphylococcus Lugdunensis
Organism: Staphylococcus lugdunensis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: PEG, EDO, OXM, FMT |
 |
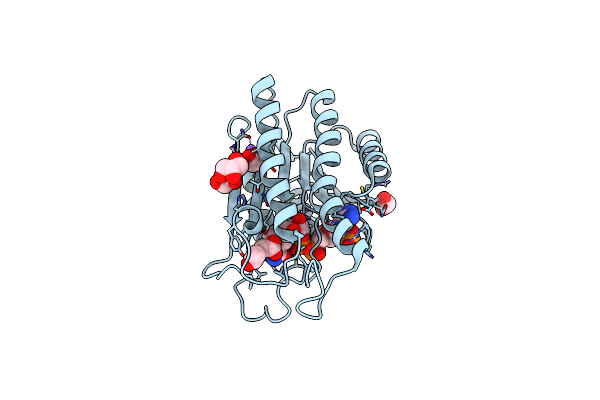
Crystal Structure Of Dye Decolorizing Peroxidase From Bacillus Subtilis At Acidic Ph
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-24 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, EDO, CIT, PEG, NA, CL, GOL |
 |
Organism: Helicoverpa armigera
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-27 Classification: OXIDOREDUCTASE Ligands: NAP, ACT, GOL, EDO, NA |
 |
 |
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: EDO, GLU, GLY, FES, FE2, GOL |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, TLA, SRT, EDO |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, PHT |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, UB7 |
 |
Crystal Structure Of Veratryl Alcohol Bound Dye Decolorizing Peroxidase From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, MPD, GOL, CL, PO4, VOH, EPE |
 |
Crystal Structure Of Hepes Bound Dye Decolorizing Peroxidase From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, EPE, GOL, NA, MPD, PO4, CL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, OXY, GOL, CL, MPD |
 |
Structure Of Cd-Bound Periplasmic Metal Binding Protein From Candidatus Liberibacter Asiaticus
Organism: Candidatus liberibacter asiaticus str. psy62
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-01-15 Classification: METAL BINDING PROTEIN Ligands: CD, GOL, SO4, ACT |
 |
Crystal Structure Of Putative Amino Acid Binding Periplasmic Abc Transporter Protein From Candidatus Liberibacter Asiaticus In Complex With Cystine
Organism: Liberibacter asiaticus (strain psy62)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-12 Classification: TRANSPORT PROTEIN Ligands: EDO, GOL, TRS, ACT, CYS |
 |
Crystal Structure Of The Putative Amino Acid-Binding Periplasmic Abc Transporter Protein From Candidatus Liberibacter Asiaticus In Complex With Cysteine
Organism: Liberibacter asiaticus (strain psy62)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-06-12 Classification: TRANSPORT PROTEIN Ligands: SO4, EDO, TRS, ACT, GOL, CYS |
 |
Crystal Structure Of Putative Amino Acid Binding Periplasmic Abc Transporter Protein From Candidatus Liberibacter Asiaticus Bound With Citrate
Organism: Liberibacter asiaticus (strain psy62)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-06-12 Classification: TRANSPORT PROTEIN Ligands: GOL, EDO, CIT, PEG |
 |
Crystal Structure Of Putative Amino Acid Binding Periplasmic Abc Transporter Protein From Candidatus Liberibacter Asiaticus In Complex With Arginine
Organism: Liberibacter asiaticus (strain psy62)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-06-12 Classification: TRANSPORT PROTEIN Ligands: EDO, SO4, ARG, ACT |

