Search Count: 32
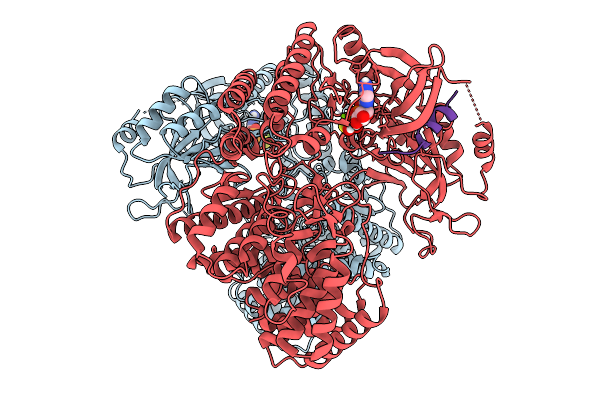 |
Cryo-Em Structure Of Classiii Salivaricin Modification Enzyme Salkc In The Presence Of Sala
Organism: Streptococcus salivarius
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: AGS, MG |
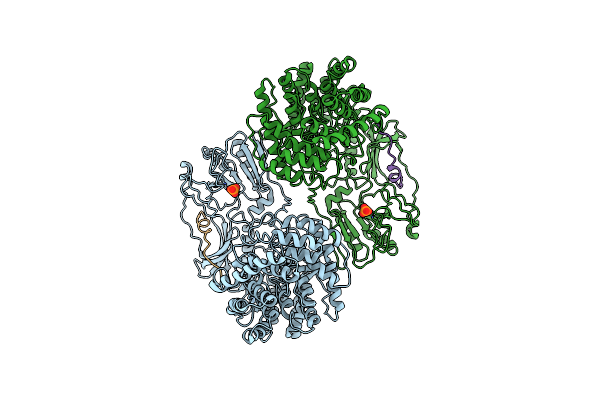 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc Mutant H522A.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: ANTIMICROBIAL PROTEIN Ligands: PO4 |
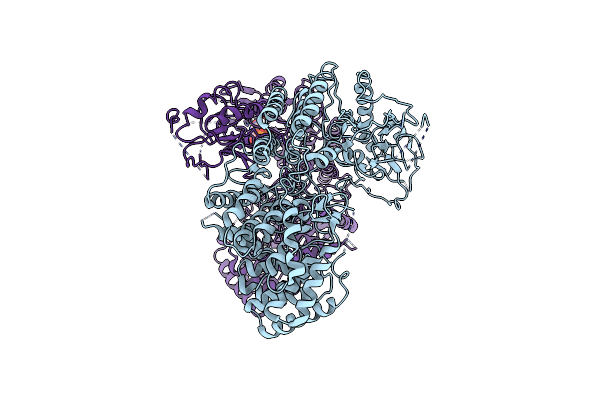 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc In The Presence Of Gtp.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: GTP |
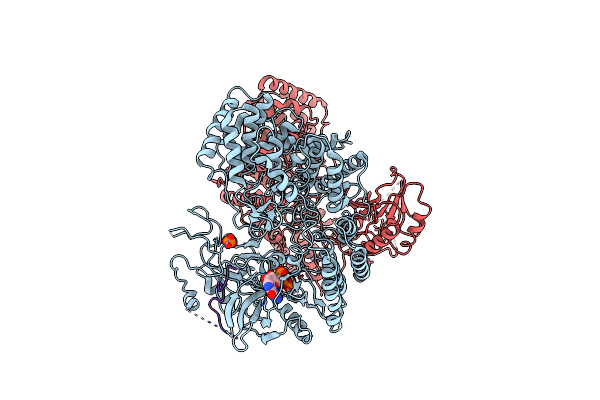 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc With Chain A Bounded To Substrate Pnea And Gtp.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: GTP, PO4 |
 |
Cryo-Em Structure Of Classiii Lanthipeptide Modification Enzyme Pnekc In The Presence Of Pnea And Gtprs.
Organism: Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: ANTIMICROBIAL PROTEIN Ligands: MG, GSP, PO4 |
 |
Organism: Streptococcus pneumoniae d39, Streptococcus streptococcus pneumoniae d39
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: HYDROLASE, TRANSLOCASE |
 |
Organism: Streptococcus pneumoniae d39
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: ATP, ZN |
 |
Organism: Streptococcus pneumoniae r6
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
Cryo-Em Structure Of Nucleotide-Bound Coma At Outward-Facing State With Ec Gate Closed Conformation
Organism: Streptococcus pneumoniae d39
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: AGS |
 |
Cryo-Em Structure Of Nucleotide-Bound Coma At Outward-Facing State With Ec Gate Open Conformation
Organism: Streptococcus pneumoniae d39
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: AGS, MG |
 |
Organism: Streptococcus pneumoniae d39
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: HYDROLASE, TRANSLOCASE Ligands: ATP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: GEC, NDP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: GEF, NDP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: GFR, NDP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: GNU, NDP |
 |
Organism: Isatis tinctoria
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: GO6, NDP |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-06-09 Classification: OXIDOREDUCTASE Ligands: NDP |

