Search Count: 27
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: GFD, A1EMI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: A1EMN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LIGASE Ligands: GFD, A1EMM |
 |
Organism: African swine fever virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-30 Classification: DNA BINDING PROTEIN Ligands: EDO, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: CHAPERONE Ligands: ATP |
 |
Crystal Structure Of The Malonyl-Acp Decarboxylase Madb From Pseudomonas Putida
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2023-03-01 Classification: TRANSFERASE |
 |
Structure Of Cdk9 In Complex With Cyclin T And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2013-04-17 Classification: TRANSFERASE/CELL CYCLE Ligands: T3C, GOL |
 |
Structure Of Cdk2 In Complex With Cyclin A And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2013-04-17 Classification: TRANSFERASE/CELL CYCLE Ligands: T3C, SO4, SGM |
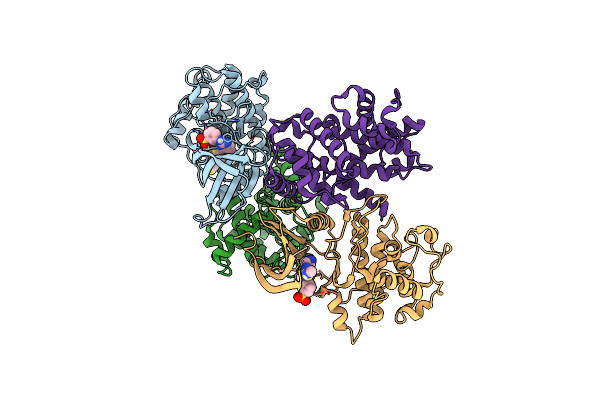 |
Structure Of Cdk2 In Complex With Cyclin A And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-03-06 Classification: TRANSFERASE/CELL CYCLE Ligands: T3E, SGM |
 |
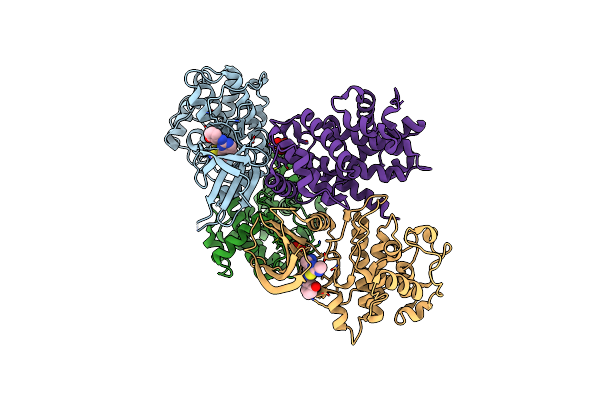
Structure Of Cdk2 In Complex With Cyclin A And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-03-06 Classification: TRANSFERASE/CELL CYCLE Ligands: T7Z, SGM |
 |
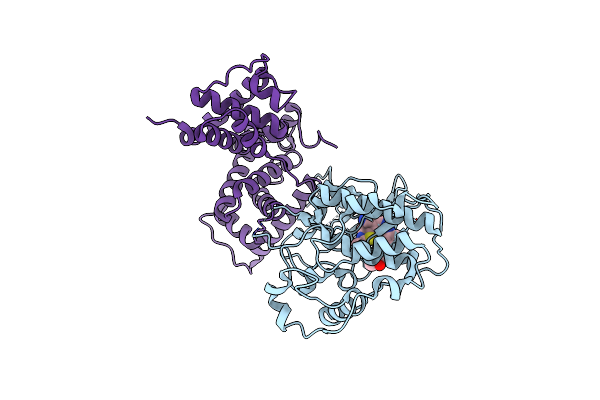
Structure Of Cdk2 In Complex With Cyclin A And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-03-06 Classification: TRANSFERASE/CELL CYCLE Ligands: T9N, SO4 |
 |
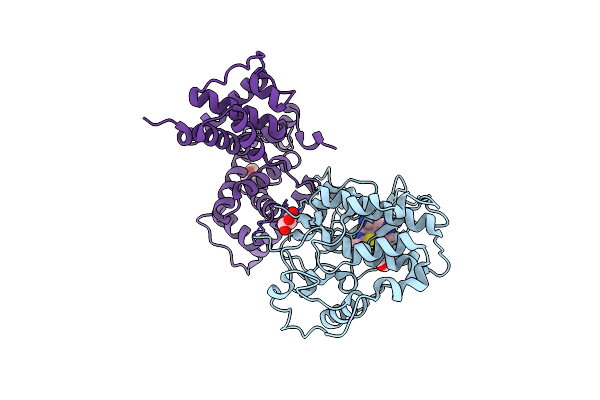
Structure Of Cdk9 In Complex With Cyclin T And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2013-01-09 Classification: TRANSFERASE/CELL CYCLE Ligands: T6Q |
 |
Structure Of Cdk9 In Complex With Cyclin T And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2013-01-09 Classification: TRANSFERASE/CELL CYCLE Ligands: T7Z, GOL |
 |
Structure Of Cdk9 In Complex With Cyclin T And A 2-Amino-4-Heteroaryl- Pyrimidine Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-01-09 Classification: TRANSFERASE/CELL CYCLE Ligands: T3E |

