Search Count: 20
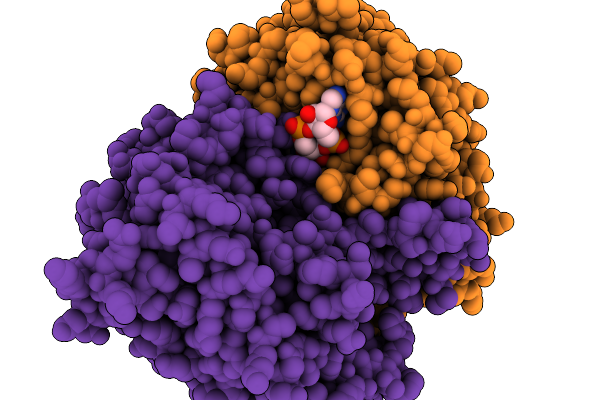 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
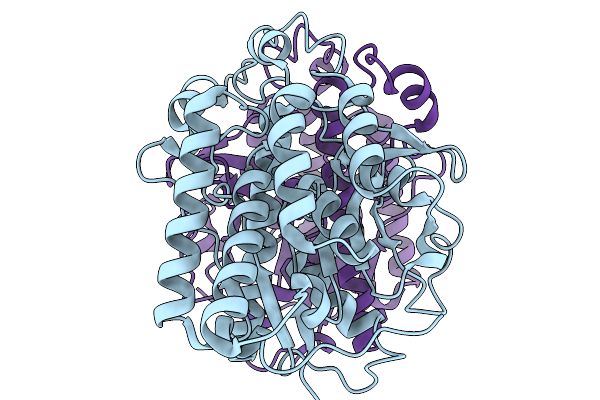 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
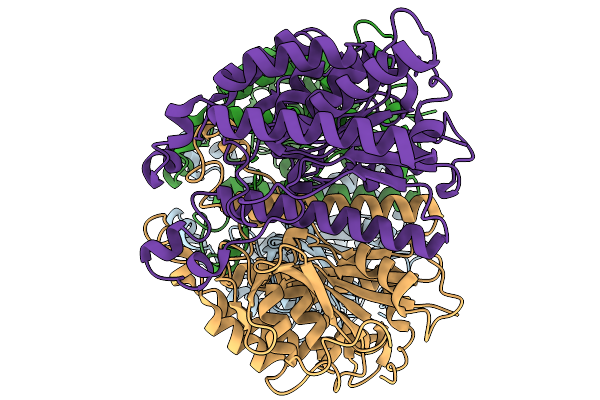 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
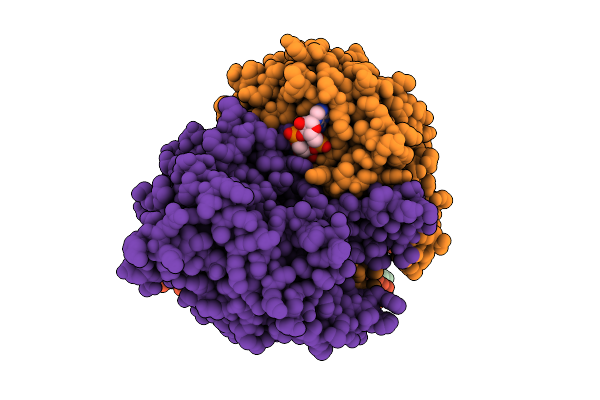 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN/DNA |
 |
Human Ogg1 Bound To A Nucleosome Core Particle With 8-Oxodguo Lesion At Shl6.0
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of P450Bsbeta-L78G/Q85F/F173S/G290I Variant In Complex With Palmitoleic Acid
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-11-08 Classification: HYDROLASE Ligands: HEM, PAM, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-03-15 Classification: IMMUNE SYSTEM Ligands: GOL, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2017-03-15 Classification: IMMUNE SYSTEM Ligands: GOL, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-15 Classification: IMMUNE SYSTEM Ligands: SO4, ACT, NAG, GOL |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-18 Classification: HYDROLASE Ligands: XQO, CA |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-18 Classification: HYDROLASE Ligands: EDO, XRJ, CA |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-18 Classification: HYDROLASE Ligands: EDO, V0N, CA |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-11-18 Classification: HYDROLASE Ligands: CL, YWN, EDO, CA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2013-05-01 Classification: HYDROLASE Ligands: CA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-01 Classification: HYDROLASE |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: GOL, NH4, NHT |
 |
Crystal Structure And Function Of Human Thioesterase Superfamily Member 2(Them2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 1998-11-04 Classification: ANTIMICROBIAL PEPTIDE |

