Search Count: 33
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Cryo-Em Structure Of Vcpip1 Vcpid Bound To Vcp D2 Domain Dimer (With Extra D2 Domain)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Cryo-Em Structure Of Vcpip1 Ubx Domain Bound To Vcp N-Domain (With D1 Domain)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: HYDROLASE |
 |
Crystal Structure Of Methanomethylophilus Alvus Pylrs(N166A/V168A) Complexed With Meta-Trifluoromethyl-2-Benzylmalonate And Amp-Pnp
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: TRANSLATION/INHIBITOR Ligands: LE0, ANP, TRS, MG |
 |
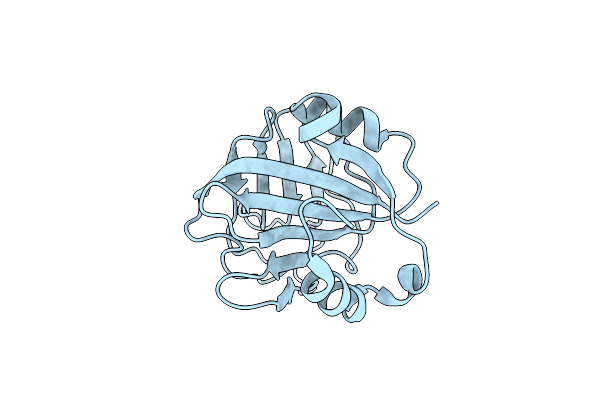
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-24 Classification: ANTIVIRAL PROTEIN |
 |
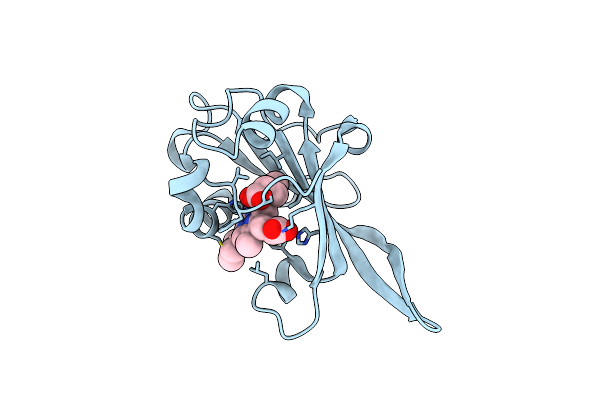
Crystal To Structure Pipeline For Ambient Temperature, In Situ Crystallography At Beamline Vmxi
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-05-24 Classification: ANTIVIRAL PROTEIN |
 |
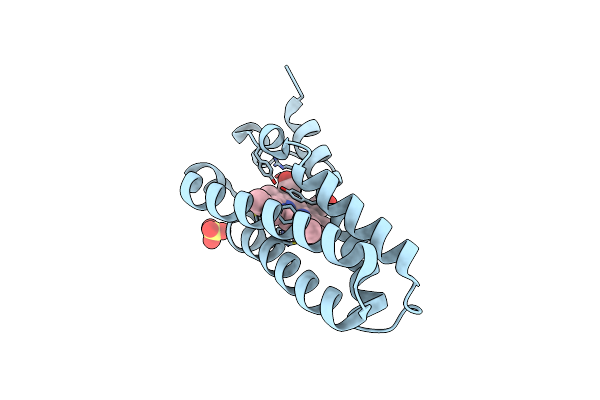
Room Temperature Crystal Structure Of Cytochrome C' From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-05-24 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
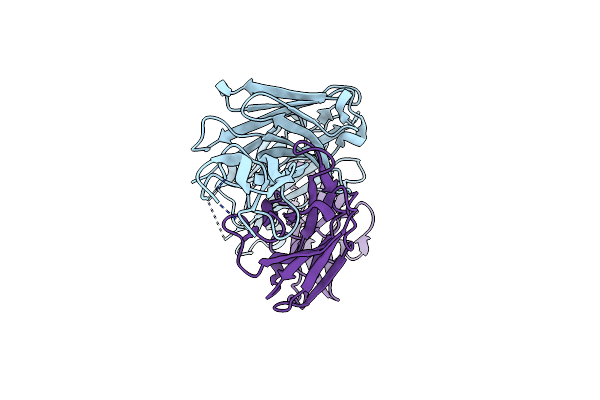
Room Temperature Crystal Structure Of Cytochrome C' From Hydrogenophilus Thermoluteolus
Organism: Hydrogenophilus thermoluteolus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-05-24 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-05-24 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of A Solute Receptor From Synechococcus Cc9311 In Complex With Alpha-Ketovaleric And Calcium
Organism: Synechococcus sp. (strain cc9311)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-04-19 Classification: TRANSPORT PROTEIN Ligands: 69O, EDO, CA |
 |
Multicrystal Room Temperature Structure Of Lysozyme Collected Using A Double Multilayer Monochromator.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-20 Classification: HYDROLASE |
 |
Room Temperature Crystal Structure Of Phnd From Synechococcus Mits9220 In Complex With Phosphate
Organism: Synechococcus sp. mit s9220
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-04 Classification: TRANSPORT PROTEIN Ligands: PO4, CL |
 |
Crystal Structure Of Phnd From Synechococcus Mits9220 In Complex With Phosphate
Organism: Synechococcus sp. mit s9220
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2021-10-27 Classification: TRANSPORT PROTEIN Ligands: CL, EDO, PO4 |
 |
Crystal Structure Of Urta1 From Synechococcus Wh8102 In Complex With Urea And Calcium
Organism: Synechococcus sp. (strain wh8102)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-10-20 Classification: TRANSPORT PROTEIN Ligands: URE, EDO, CA |
 |
Crystal Structure Of Urta From Synechococcus Cc9311 In Complex With Urea And Calcium
Organism: Synechococcus sp. (strain cc9311)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-10-13 Classification: TRANSPORT PROTEIN Ligands: URE, EDO, CA, CL |
 |
Crystal Structure Of A Putative Oligosaccharide Periplasmic-Binding Protein From Synechococcus Sp. Mits9220 In Complex With Zinc
Organism: Synechococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2021-05-12 Classification: TRANSPORT PROTEIN Ligands: GOL, EDO, ZN, SO4 |
 |
Crystal Structure Of A Putative Oligosaccharide Periplasmic-Binding Protein From Synechococcus Sp. Mits9220
Organism: Synechococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2021-05-12 Classification: TRANSPORT PROTEIN Ligands: EDO, GOL |

