Search Count: 26
 |
Dpo4 Dna Polymerase (Wild Type) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus p2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: CA, DTP |
 |
Dpo4 Dna Polymerase (R332A) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: DTP, CA, PO4 |
 |
Dpo4 Dna Polymerase (R336A) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: DTP, CA, PO4 |
 |
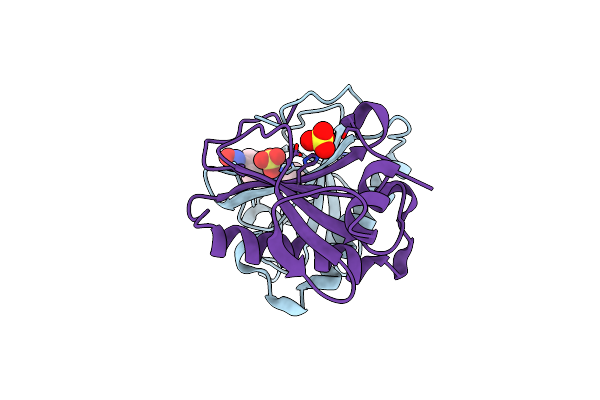
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With Kv24
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: A1IQU, SO4 |
 |
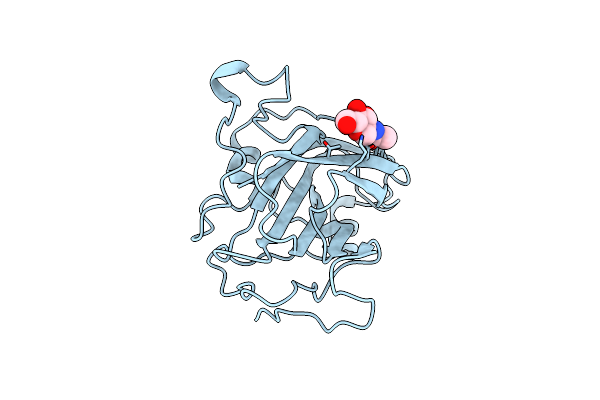
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With Kv30
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: A1IQV, SO4 |
 |
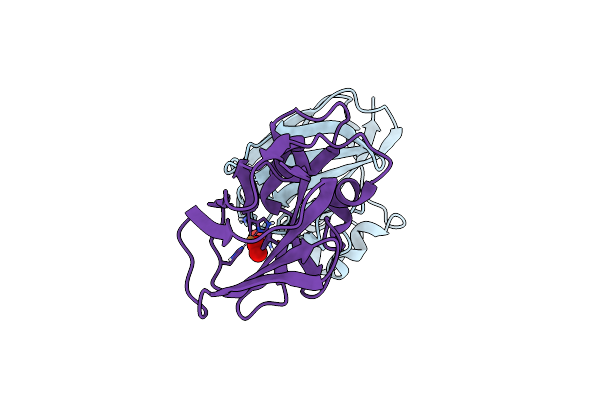
Organism: Neosartorya fumigata (strain cea10 / cbs 144.89 / fgsc a1163)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAG |
 |
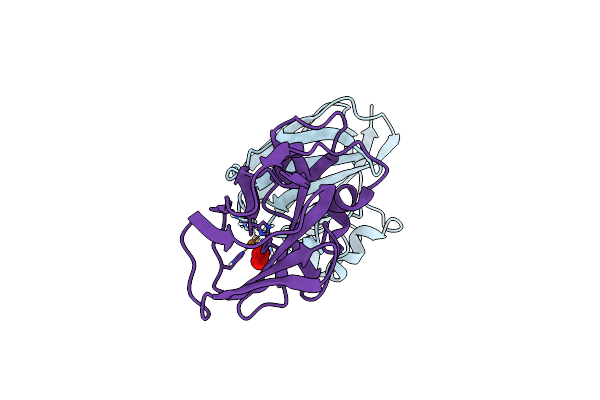
Room Temperature X-Ray Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase At 1.1 Angstrom Resolution.
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Neutron Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Crystal Structure Of The Saccharomyces Cerevisiae Rtr1 (Regulator Of Transcription)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-05-04 Classification: HYDROLASE Ligands: ZN, SO4, GOL |
 |
Structural And Functional Analysis Of A Lytic Polysaccharide Monooxygenase Important For Efficient Utilization Of Chitin In Cellvibrio Japonicus
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-02-17 Classification: SUGAR BINDING PROTEIN Ligands: CU |
 |
Structural And Functional Characterization Of A Chitin-Active 15.5 Kda Lytic Polysaccharide Monooxygenase Domain From A Modular Chitinase From Jonesia Denitrificans
Organism: Jonesia denitrificans
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-12-09 Classification: HYDROLASE Ligands: CU1 |
 |
Crystal Structure Of Dyrk1A In Complex With 10-Iodo-Substituted 11H-Indolo[3,2-C]Quinoline-6-Carboxylic Acid Inhibitor 5J
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2015-03-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: PG4, SO4, 4E1 |
 |
Crystal Structure Of Dyrk1A In Complex With 10-Chloro-Substituted 11H-Indolo[3,2-C]Quinolone-6-Carboxylic Acid Inhibitor 5S
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-03-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, EDO, 4E2, IOD |
 |
Crystal Structure Of Dyrk1Aa In Complex With 10-Bromo-Substituted 11H-Indolo[3,2-C]Quinolone-6-Carboxylic Acid Inhibitor 5T
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-03-25 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, EDO, 4E3 |
 |
Novel Modifications On C-Terminal Domain Of Rna Polymerase Ii Can Fine- Tune The Phosphatase Activity Of Ssu72.
Organism: Drosophila melanogaster, Synthetic
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-08-07 Classification: HYDROLASE Ligands: PO4 |
 |
Novel Modifications On C-Terminal Domain Of Rna Polymerase Ii Can Fine-Tune The Phosphatase Activity Of Ssu72
Organism: Drosophila melanogaster, Synthetic
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-08-07 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-10 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-09-26 Classification: HYDROLASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-07-18 Classification: RIBOSOME Ligands: MG, K, AM2, ZN |
 |
Crystal Structure Of Reductase (R) Domain Of Non-Ribosomal Peptide Synthetase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-06-20 Classification: LIGASE |

