Search Count: 54
 |
Cyanide Dihydratase From Bacillus Pumilus C1 E155R Variant With Altered Helical Twist.
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: HYDROLASE |
 |
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: HYDROLASE |
 |
A Mutant Of The Nitrile Hydratase From Geobacillus Pallidus Having Enhanced Thermostability
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-29 Classification: LYASE Ligands: CO, CL |
 |
A Mutant Of The Nitrile Hydratase From Geobacillus Pallidus Having Enhanced Thermostability
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-03-08 Classification: LYASE Ligands: CO, CL |
 |
A Mutant Of The Nitrile Hydratase From Geobacillus Pallidus Having Enhanced Thermostability
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-01-18 Classification: LYASE Ligands: CO, MG, CL |
 |
A Mutant Of The Nitrile Hydratase From Geobacillus Pallidus Having Enhanced Thermostability
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-01-18 Classification: LYASE Ligands: CO, MG, CL |
 |
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-01-18 Classification: LYASE Ligands: 3CO, CL |
 |
A Mutant Of The Nitrile Hydratase From Geobacillus Pallidus Having Enhanced Thermostability
Organism: Aeribacillus pallidus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-01-18 Classification: LYASE Ligands: 3CO, CL |
 |
Cyanide Dihydratase From Bacillus Pumilus C1 Variant - Q86R,H305K,H308K,H323K
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: HYDROLASE |
 |
Structure Of Full-Length, Monomeric, Soluble Somatic Angiotensin I-Converting Enzyme Showing The N- And C-Terminal Ellipsoid Domains
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: ZN, CL, NAG |
 |
Local Refinement Structure Of The N-Domain Of Full-Length, Monomeric, Soluble Somatic Angiotensin I-Converting Enzyme
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: ZN, NAG |
 |
Local Refinement Structure Of The C-Domain Of Full-Length, Monomeric, Soluble Somatic Angiotensin I-Converting Enzyme
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: CL, NAG |
 |
Local Refinement Structure Of The Two Interacting N-Domains Of Full-Length, Dimeric, Soluble Somatic Angiotensin I-Converting Enzyme
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: ZN, NAG |
 |
Local Refinement Structure Of A Single N-Domain Of Full-Length, Dimeric, Soluble Somatic Angiotensin I-Converting Enzyme
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: HYDROLASE Ligands: ZN, NAG |
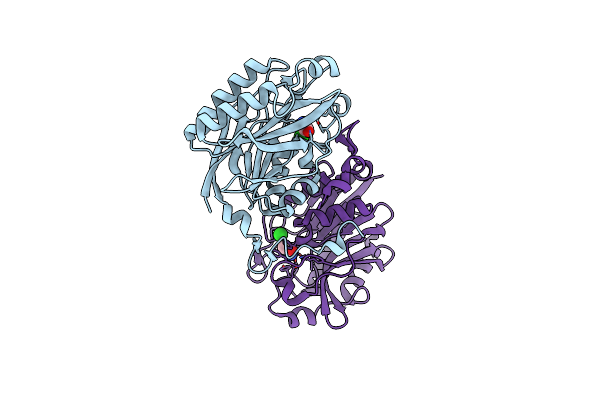 |
The C146A Variant Of An Amidase From Pyrococcus Horikoshii With Bound Acetamide
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: ACM, CL |
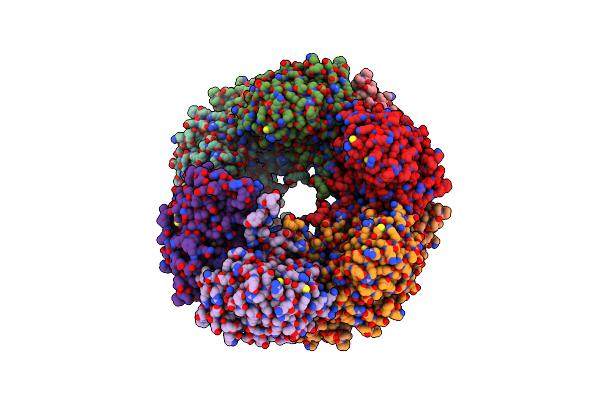 |
Cryo-Em Structure Of The Nitrilase From Pseudomonas Fluorescens Ebc191 At 3.3 Angstroms
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: HYDROLASE |
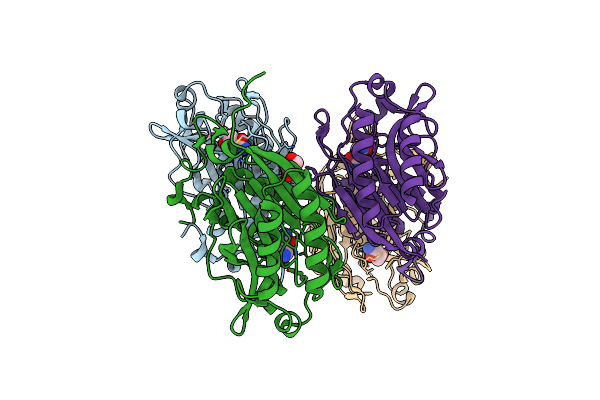 |
The C146A Variant Of An Amidase From Pyrococcus Horikoshii With Bound Glutaramide
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: P6W, GOL |
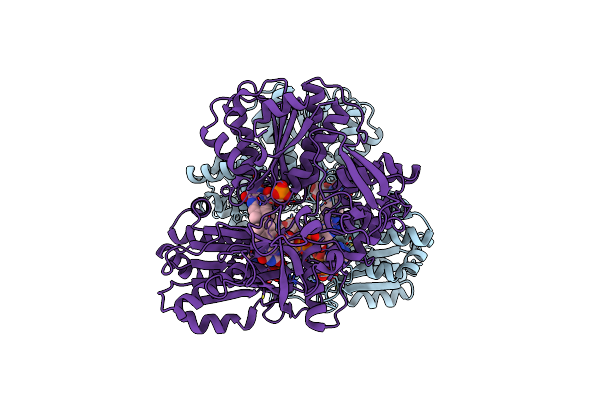 |
Organism: Candida tropicalis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2020-01-08 Classification: OXIDOREDUCTASE Ligands: FAD, FMN, NDP |
 |
Organism: Candida tropicalis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-08 Classification: OXIDOREDUCTASE Ligands: FAD, FMN |
 |
Organism: Nesterenkonia sp. 10004
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-06-20 Classification: HYDROLASE |

