Search Count: 15
 |
Organism: Geobacter metallireducens (strain atcc 53774 / dsm 7210 / gs-15)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Geobacter metallireducens (strain atcc 53774 / dsm 7210 / gs-15)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: ZN, PE8, PO4, MG, COA, CL, EPE |
 |
Organism: Geobacter metallireducens gs-15
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: ZN, PE8, PO4, MG, COA, CL, EPE |
 |
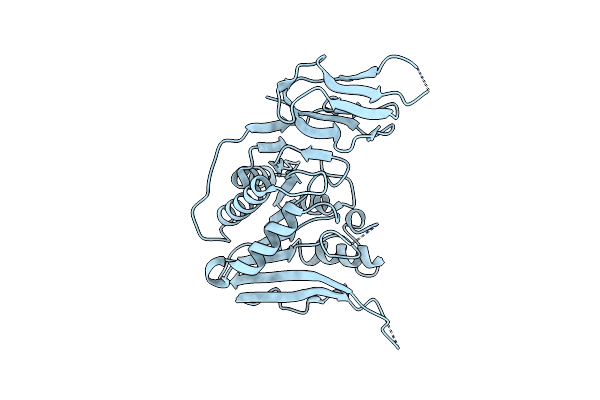
Apo Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-20 Classification: HYDROLASE |
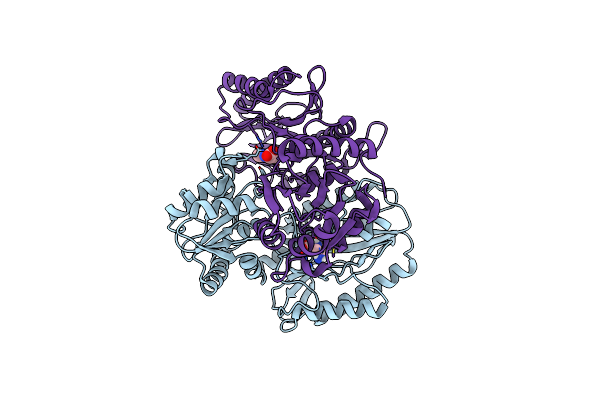 |
Substrate Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: P4B, 4CS |
 |
Apo Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-20 Classification: HYDROLASE |
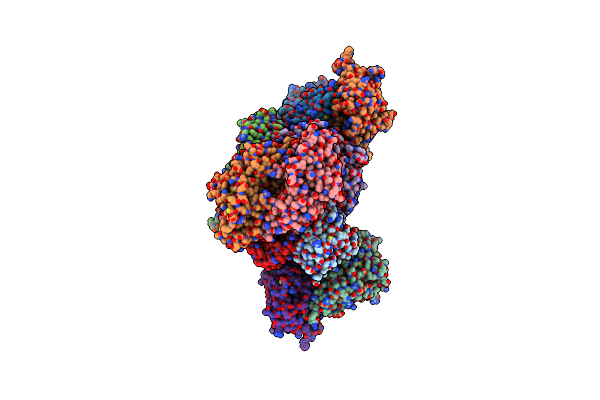 |
Product Bound Structure Of The Ectoine Utilization Protein Eute (Doeb) From Ruegeria Pomeroyi
Organism: Ruegeria pomeroyi (strain atcc 700808 / dsm 15171 / dss-3)
Method: X-RAY DIFFRACTION Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, DAB, ACT |
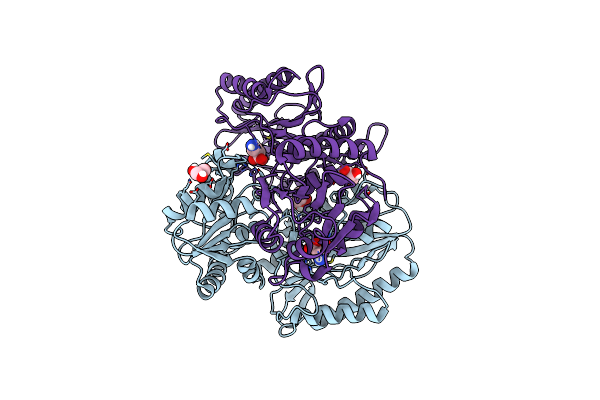 |
Product Bound Structure Of The Ectoine Utilization Protein Eutd (Doea) From Halomonas Elongata
Organism: Halomonas elongata
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, P4B |
 |
Organism: Paenibacillus lautus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2018-08-22 Classification: METAL BINDING PROTEIN Ligands: FE, SO4 |
 |
Organism: Paenibacillus lautus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-08-22 Classification: METAL BINDING PROTEIN Ligands: FE, 9YT |
 |
Organism: Paenibacillus lautus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-22 Classification: METAL BINDING PROTEIN Ligands: FE, 4CS |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-28 Classification: TRANSPORT PROTEIN Ligands: 3Q7 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-03-14 Classification: CHOLINE-BINDING PROTEIN Ligands: 1Y8, EDO |
 |
Crystal Structure Of The Ectoine Synthase From The Cold-Adapted Marine Bacterium Sphingopyxis Alaskensis
Organism: Sphingopyxis alaskensis rb2256
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-27 Classification: LYASE |
 |
High Resolution Structure Of The Ectoine Synthase From The Cold-Adapted Marine Bacterium Sphingopyxis Alaskensis
Organism: Sphingopyxis alaskensis (strain dsm 13593 / lmg 18877 / rb2256)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-04-27 Classification: LYASE Ligands: PGO |

