Search Count: 25
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.29 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: NAD, EDO |
 |
Crystal Structure Of A Chimeric Alpha-Amylase From Pseudoalteromonas Haloplanktis Complexed With Rearranged Acarbose
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: CL, CA, EDO |
 |
Crystal Structure Of A Chimeric Alpha-Amylase From Pseudoalteromonas Haloplanktis
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-06-21 Classification: HYDROLASE Ligands: EDO, CL, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: U9Y |
 |
Vim-2_1Dj- Triazole Inhibitors With Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-02 Classification: ANTIBIOTIC Ligands: ZN, OH, CL, 9O8, IOD |
 |
Vim-2_1Di-Triazole Inhibitors With Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-09-02 Classification: ANTIBIOTIC Ligands: ZN, OH, CL, 9OB, IOD |
 |
Vim-2_1Cc-. Triazole Inhibitors With Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-07-22 Classification: ANTIBIOTIC Ligands: 9NW, ZN, OH |
 |
Vim-2_1Dh-Triazole Inhibitors With Promising Inhibitor Effects Against Antibiotic Resistance Metallo-Beta-Lactamases
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-07-22 Classification: ANTIBIOTIC Ligands: NL5, ZN, OH, IOD |
 |
Crystal Structure Of 6-Phosphogluconate Dehydrogenase With ((4R,5R)-5-(Hydroxycarbamoyl)-2,2-Dimethyl-1,3-Dioxolan-4-Yl)Methyl Dihydrogen Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-08-22 Classification: OXIDOREDUCTASE Ligands: 8HS |
 |
Vim-2_10B. Metallo-Beta-Lactamase Inhibitors By Bioisosteric Replacement: Preparation, Activity And Binding
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: ZN, 8XZ, CL, PO4, SO4, PEG, MG |
 |
Vim-2_10C. Metallo-Beta-Lactamase Inhibitors By Bioisosteric Replacement: Preparation, Activity And Binding
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: ZN, 8XW |
 |
Vim-2_2B. Metallo-Beta-Lactamase Inhibitors By Bioisosteric Replacement: Preparation, Activity And Binding
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-03-29 Classification: HYDROLASE Ligands: ZN, 8SH, MG |
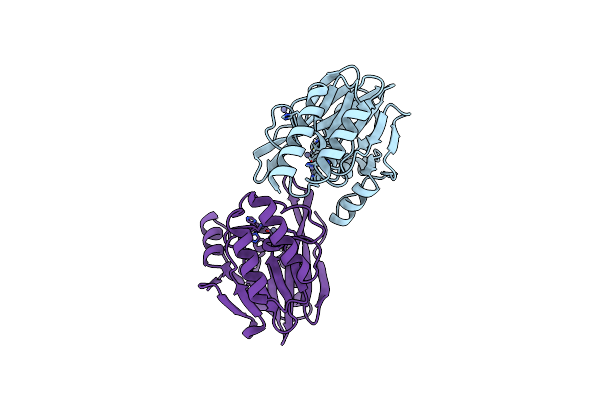 |
Tmb-1. Structural Insights Into Tmb-1 And The Role Of Residue 119 And 228 In Substrate And Inhibitor Binding
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: HYDROLASE Ligands: ZN, CL |
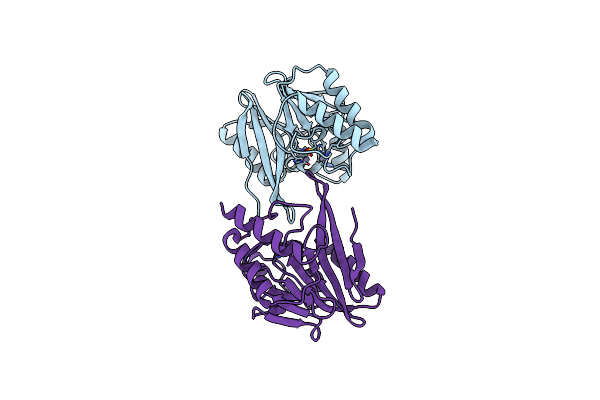 |
W228R-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN, MG |
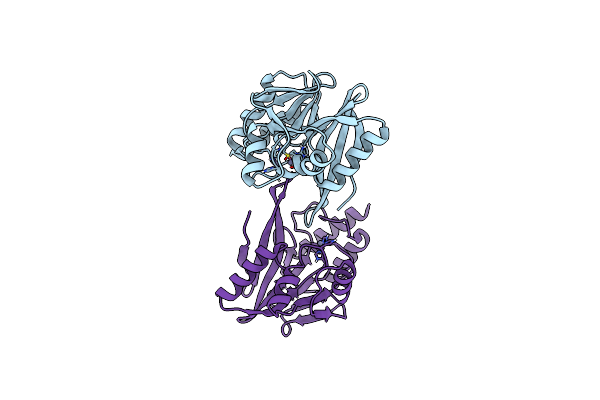 |
W228A-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
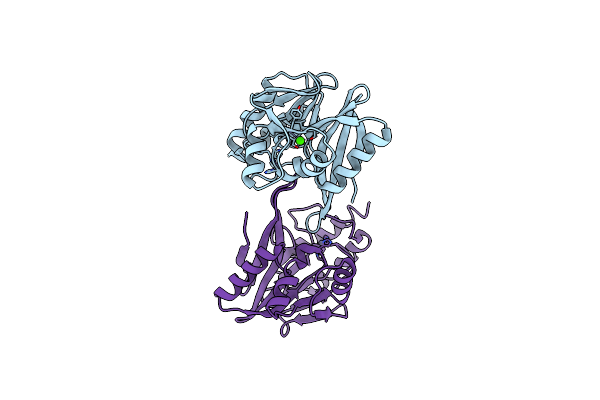 |
W228Y-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN, CA |
 |
Y233A-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
 |
W228S-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN |

