Search Count: 25
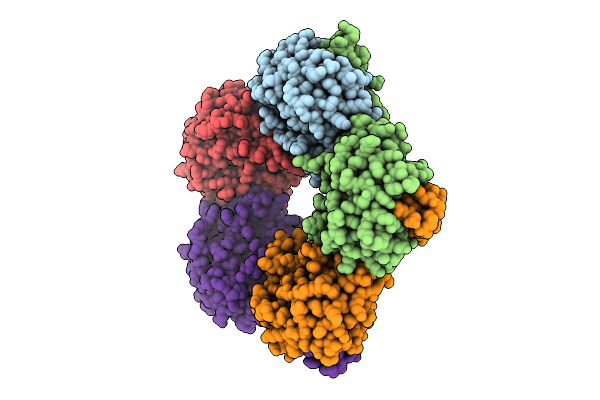 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Aeromonas sp. asnih1
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
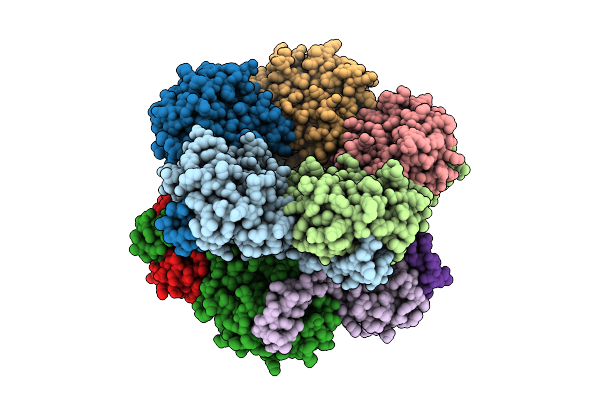 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Homogentisate 1,2-Dioxygenase From Acinetobacter In Complex With Zn Ion
Organism: Acinetobacter
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: ZN, TRS |
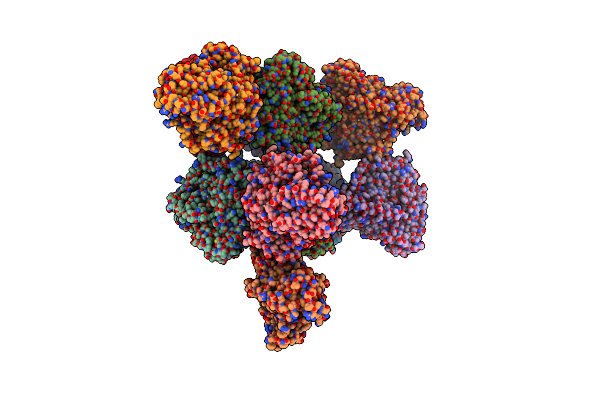 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
 |
Crystal Structure Of Escherichia Coli Glyoxylate Carboligase Double Mutant In Complex With Glycolaldehyde
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: MG, FAD, TPP, DW3, UQ1 |
 |
Crystal Structure Of Escherichia Coli Glyoxylate Carboligase Quadruple Mutant
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-11-22 Classification: LIGASE Ligands: FAD, TPP, MG, UQ0 |
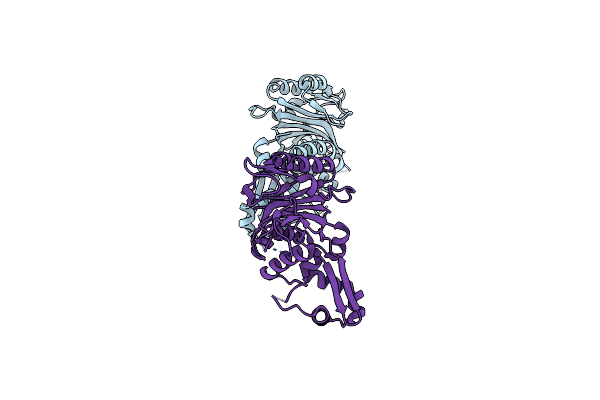 |
Crystal Structure Of The Type I-B Crispr-Associated Protein, Csh2 From Thermobaculum Terrenum
Organism: Thermobaculum terrenum
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-05-31 Classification: TRANSFERASE |
 |
Crystal Structure Of Class Ii Pyruvate Aldolase From Pseudomonas Aeruginosa.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2022-08-24 Classification: LYASE Ligands: IOD, P6G |
 |
Crystal Structure Of The Intact Mtase From Vibrio Vulnificus Yj016 In Complex With The Dna-Mimicking Ocr Protein And The S-Adenosyl-L-Homocysteine (Sah)
Organism: Vibrio vulnificus (strain yj016), Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Deinococcus metallilatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-08-25 Classification: LIGASE |
 |
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: P6G, MG, TPP, CA |
 |
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: P6G, CA, MG, TPP, DW3 |
 |
Crystal Structure Of Udp-N-Acetylmuramic Acid L-Alanine Ligase (Murc) From Mycobacterium Bovis
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-22 Classification: LIGASE Ligands: ZN |
 |
Crystal Structure Of Udp-N-Acetylmuramic Acid L-Alanine Ligase (Murc) From Mycobacterium Bovis In Complex With Udp-N-Acetylglucosamine
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2020-04-22 Classification: LIGASE Ligands: UD1, ZN |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Blue Fluorescent Protein From Metagenomic Library In Complex With Nadph
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
Organism: Parvibaculum lavamentivorans (strain ds-1 / dsm 13023 / ncimb 13966)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-04-10 Classification: OXIDOREDUCTASE Ligands: FAD, MG |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 10988 / dsm 424 / lmg 404 / ncimb 8938 / nrrl b-806 / zm1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-18 Classification: RNA BINDING PROTEIN |

