Search Count: 171
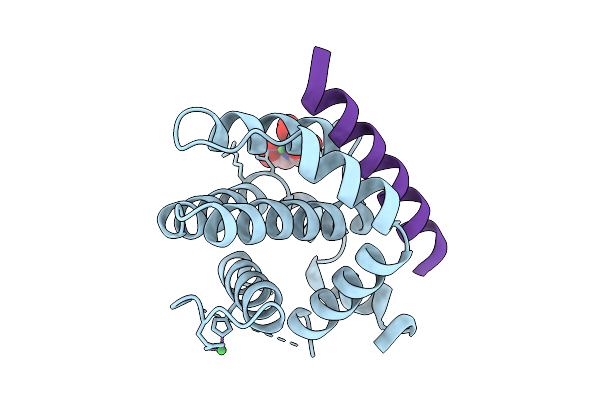 |
Crystal Structure Of Bcl-2 In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: APOPTOSIS Ligands: NI, NTA |
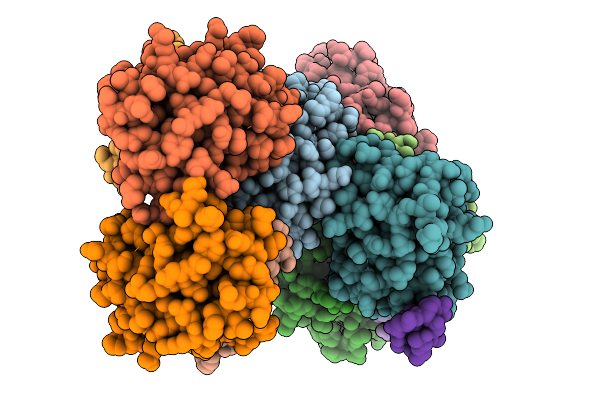 |
Crystal Structure Of Bcl-2 (G101V) Mutant In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
|
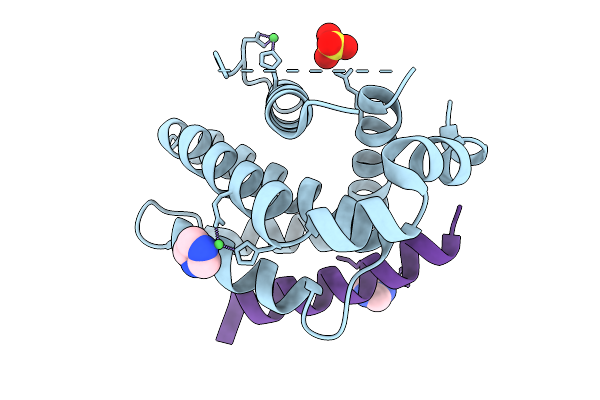 |
Crystal Structure Of Human Bcl-2 (R129L) Mutant In Complex With A Stapled Bad Bh3 Peptide Bad Sahb 4.2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: APOPTOSIS Ligands: IMD, SO4, NI |
 |
Organism: Homo sapiens, Human gammaherpesvirus 8, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRAL PROTEIN/SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE/INHIBITOR Ligands: A1AEO, SO4 |
 |
Organism: Human gammaherpesvirus 8
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRAL PROTEIN |
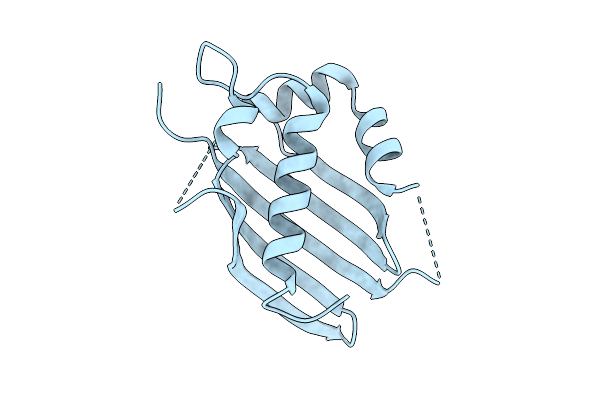 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: PROTEIN BINDING Ligands: HOH |
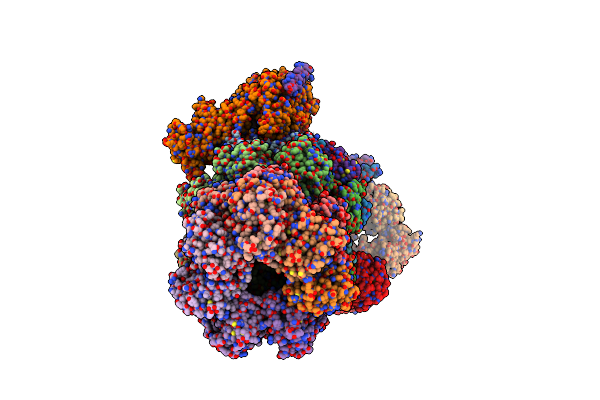 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: NUCLEAR PROTEIN Ligands: ATP, MG, ADP |
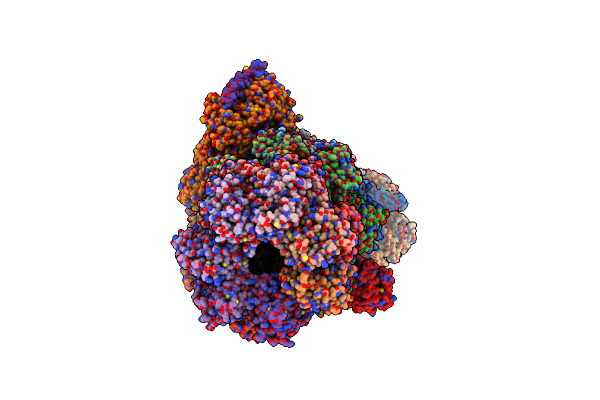 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: NUCLEAR PROTEIN Ligands: ZN, ATP, MG, ADP |
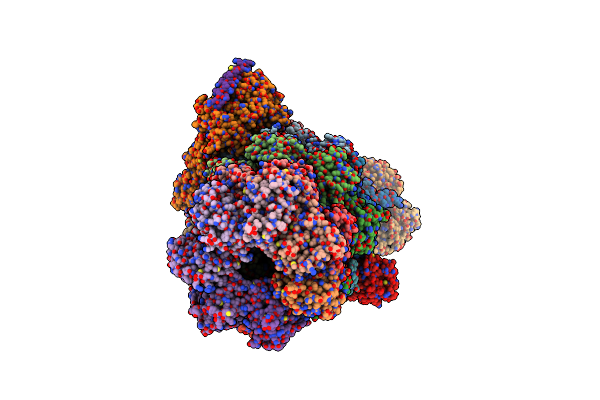 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: NUCLEAR PROTEIN Ligands: ZN, ATP, ADP |
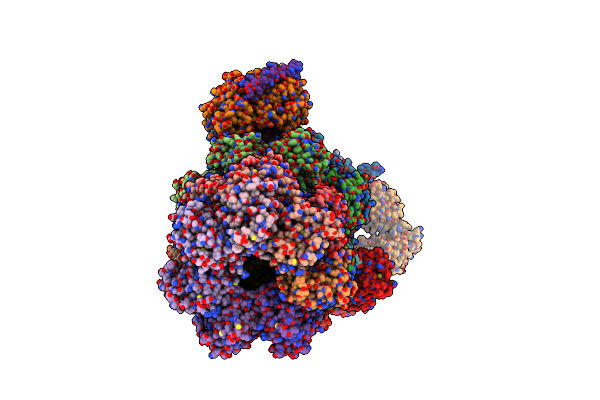 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: NUCLEAR PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: EDO, GOL |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: ZN, 1PE, ACT |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-588
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BHY, ZN, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-600
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BHX, ZN, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-584
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: ZN, A1BHW, K |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With To-589
Organism: Danio rerio
Method: X-RAY DIFFRACTION Release Date: 2024-12-04 Classification: METAL BINDING PROTEIN Ligands: A1BH8, ZN, K |

