Search Count: 44
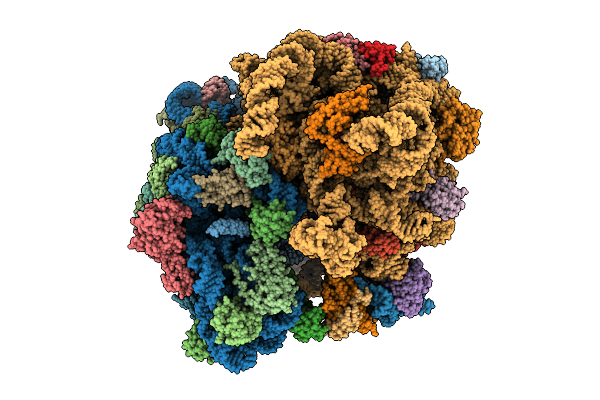 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: PHE, GNP, MG, ZN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: RIBOSOME Ligands: GNP, MG, ZN |
 |
Cryo-Em Map Of 30S Ribosomal Subunit In Complex With Metap1C Of Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: RIBOSOME Ligands: GNP |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: RIBOSOME Ligands: ERY |
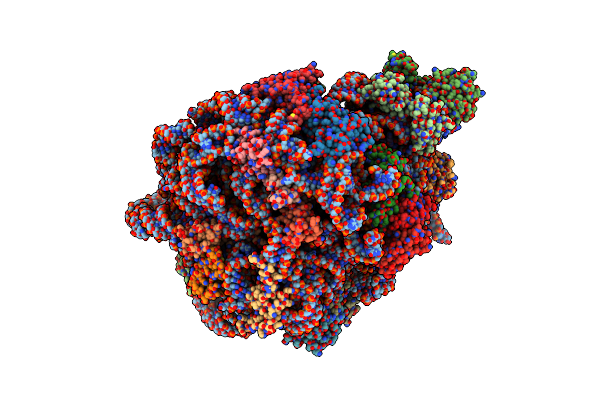 |
Mycobacterium Smegmatis 50S Ribosomal Subunit From Stationary Phase Of Growth
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2023-05-03 Classification: RIBOSOME Ligands: MG, ZN |
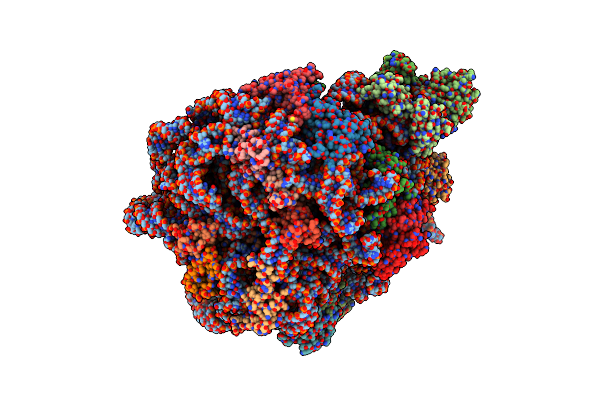 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:4.10 Å Release Date: 2023-05-03 Classification: RIBOSOME Ligands: MG, ZN |
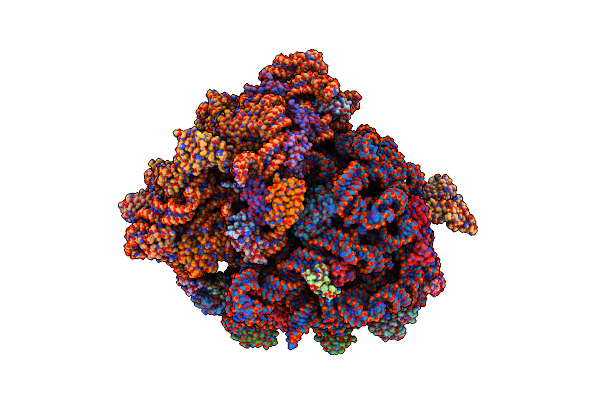 |
Molecular Model Of The Cryo-Em Structure Of 70S Ribosome In Complex With Peptide Deformylase And Trigger Factor
Organism: Escherichia coli (strain k12), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-04-07 Classification: RIBOSOME |
 |
Molecular Model Of The Cryo-Em Structure Of 70S Ribosome In Complex With Peptide Deformylase, Trigger Factor, And Methionine Aminopeptidase
Organism: Escherichia coli (strain k12), Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:4.10 Å Release Date: 2021-04-07 Classification: RIBOSOME |
 |
Crystal Structure Of Insulin Hexamer Fitted Into Cryo Em Density Map Where Each Dimer Was Kept As Rigid Body
|
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:9.80 Å Release Date: 2019-06-19 Classification: TRANSCRIPTION |
 |
E. Coli Peptide Deformylase Crystal Structure Fitted Into The Cryo-Em Density Map Of E. Coli 70S Ribosome In Complex With Peptide Deformylase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
E. Coli Methionine Aminopeptidase Crystal Structure Fitted Into The Cryo-Em Density Map Of E. Coli 70S Ribosome In Complex With Methionine Aminopeptidase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Peptide Deformylase And Methionine Aminopeptidase Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Methionine Aminopeptidase Enzyme And Chaperone Trigger Factor Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli k-12, Thermotoga maritima msb8
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Crystal Structure Of E. Coli Peptide Deformylase Enzyme And Chaperone Trigger Factor Fitted Into The Cryo-Em Density Map Of The Complex
Organism: Escherichia coli h591
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: RIBOSOME |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:10.70 Å Release Date: 2019-01-23 Classification: TRANSCRIPTION |
 |
E. Coli 50S Subunit Bound Hflx Protein In Presence Of Atp (Amp-Pnp) And Gtp (Gmp-Pnp) Analogs.
Organism: Escherichia coli (strain k12), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-06-27 Classification: RIBOSOME |
 |
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Ef-G.Gtp State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Initiation-Like State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |

