Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: BO3 |
 |
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
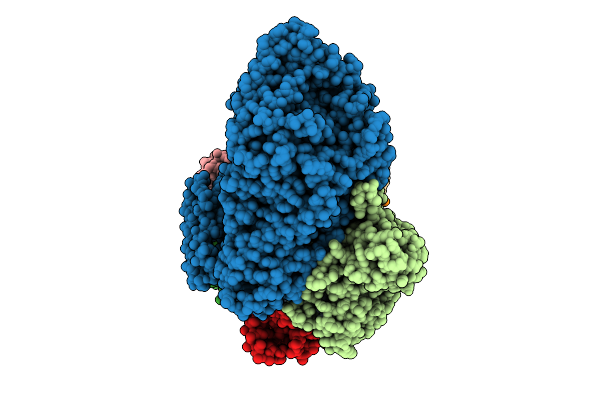
The Cryo-Em Structure Of Porcine Serum Mgam Bound With Acarviosyl-Maltotriose.
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
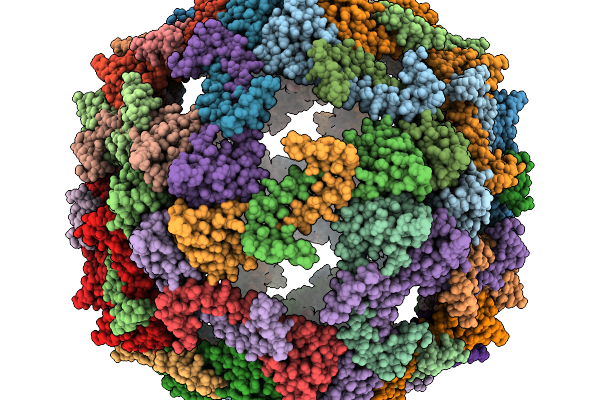
Organism: Archaeoglobus fulgidus dsm 4304, Pyrococcus furiosus dsm 3638, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
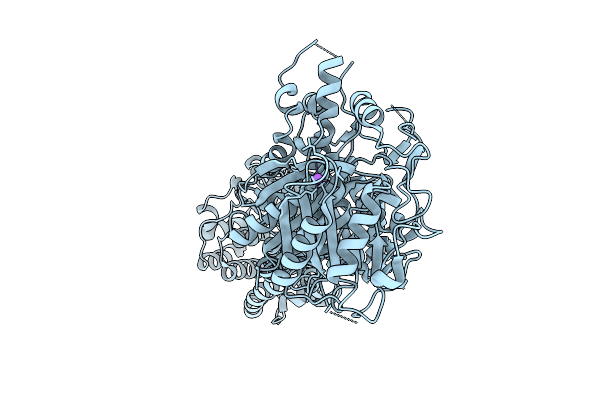
Cryo-Em Structure Of Pyrene-Modified Tip60 Double Mutant (G12C/S50C) With Addition Of Nile Red
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: A1L9F |
 |
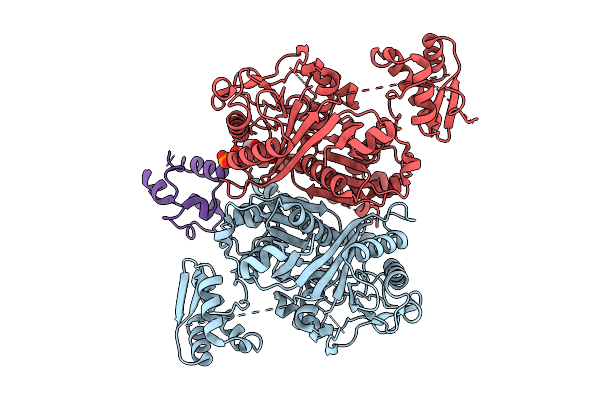
Organism: Streptomyces graminofaciens, Synthetic construt
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE |
 |
Class 3 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
 |
Class 1 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: ADP, MG |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of The Inhibitor-Bound Vo Complex From Enterococcus Hirae
Organism: Enterococcus hirae atcc 9790
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CDL, NA, W3K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN |
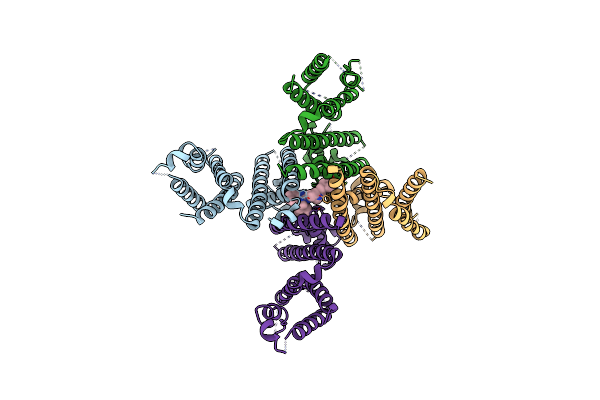 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: XB7 |
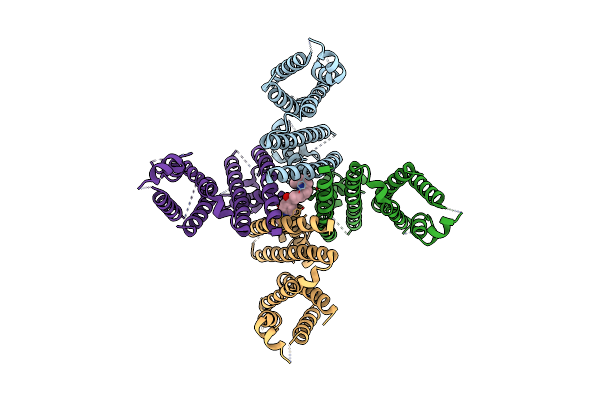 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: A1L2J |
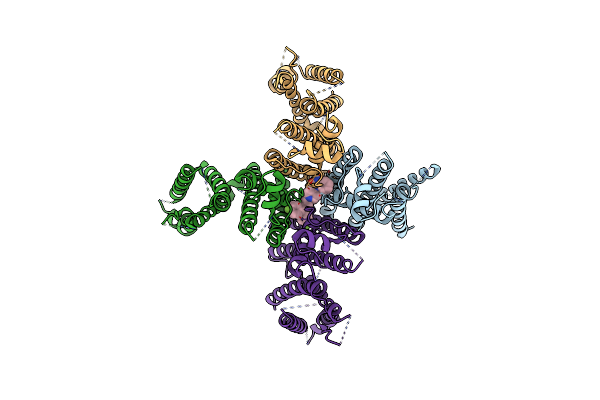 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: TRANSPORT PROTEIN Ligands: 1II |
 |
Cryo-Em Structure Of Aquifex Aeolicus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE-RNA COMPLEX |
 |
Cryo-Em Structure Of Hydrogenobacter Thermophilus Minimal Protein-Only Rnase P (Harp) In Complex With Pre-Trnas
Organism: Hydrogenobacter thermophilus dsm 653, Aquifex aeolicus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: HYDROLASE/RNA Ligands: MG |
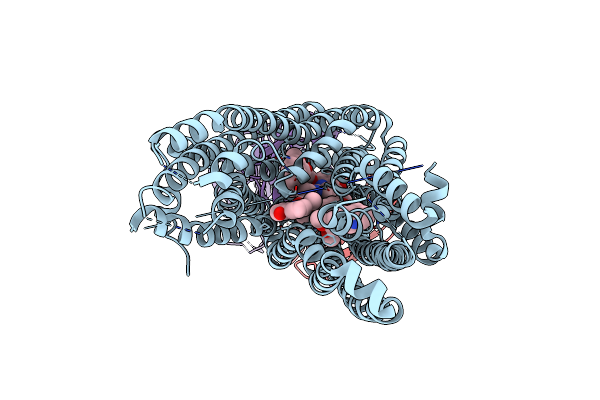 |
P-Glycoprotein In Complex With Uic2 Fab And Triple Elacridar Molecules In Lmng Detergent
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: R0Z |
 |
P-Glycoprotein In Complex With Uic2 Fab And Triple Elacridar Molecules In Nanodisc
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CLR, 3PE, R0Z |
 |
Crystal Structure Of Pyruvic Oxime Dioxygenase (Pod) From Bradyrhizobium Sp. Wsm3983
Organism: Bradyrhizobium sp. wsm3983
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: NI, SO4 |