Search Count: 901
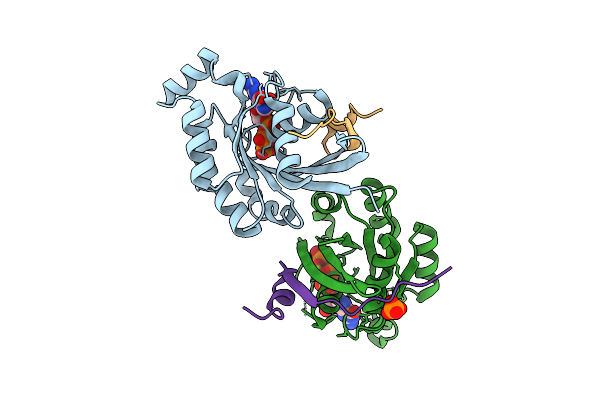 |
Crystal Structure Of Human Rac1 In Complex With The Scaffold Protein Posh (Residues 321-348)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: GNP, MG, PO4, CL |
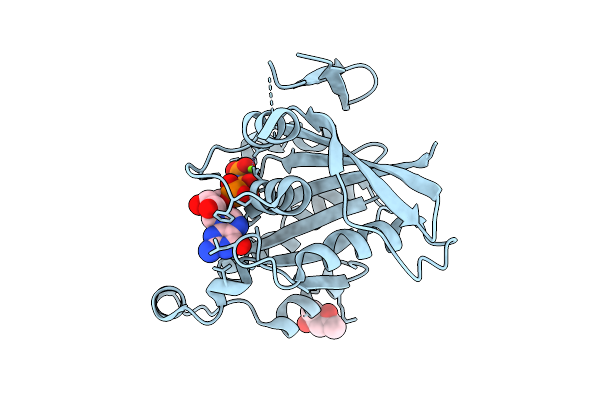 |
Crystal Structure Of Human Rac1 Fused With The Scaffold Protein Posh (Residues 319-371)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: GNP, MG, MPD |
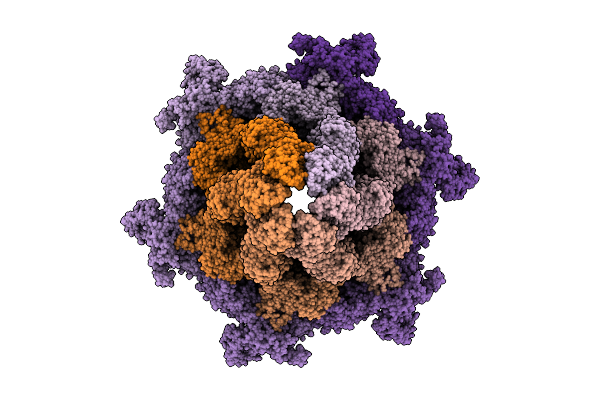 |
Cryo-Em Structure Of The Plpvc1 Baseplate, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
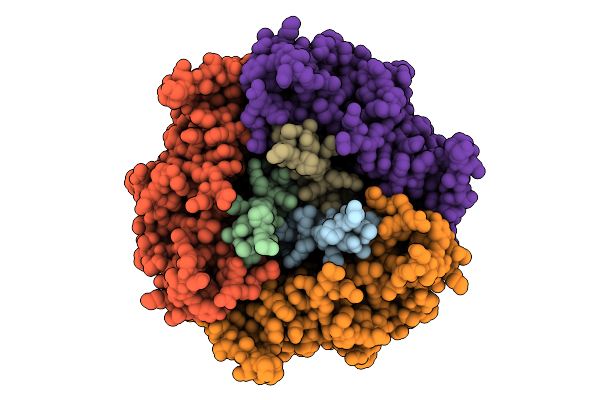 |
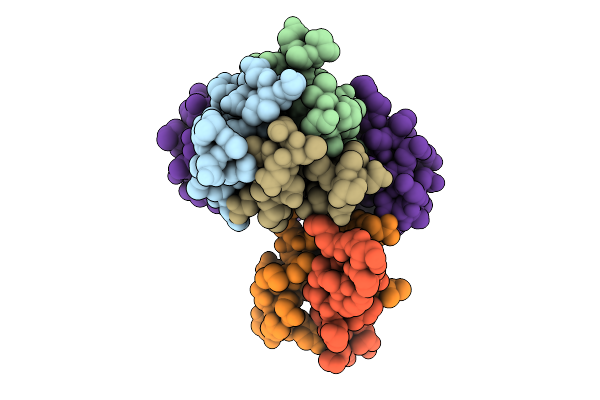
Cryo-Em Structure Of The Plpvc1 Central Spike, 3-Fold Symmetrized (C3), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
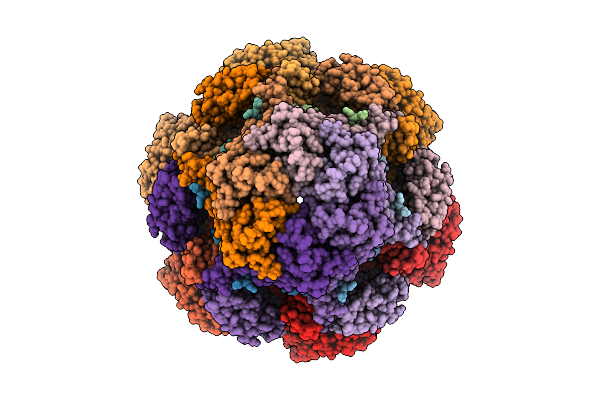 |
Cryo-Em Structure Of The Plpvc1 Cap, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
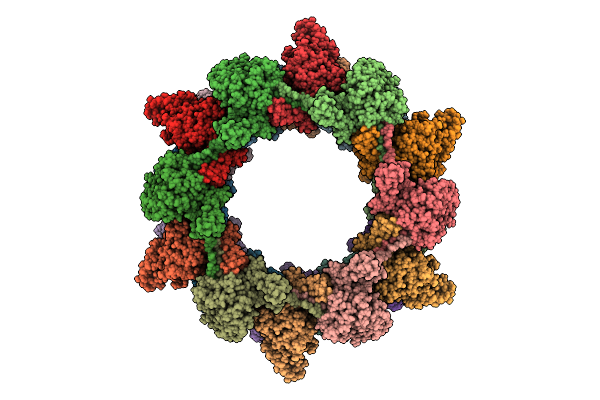
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
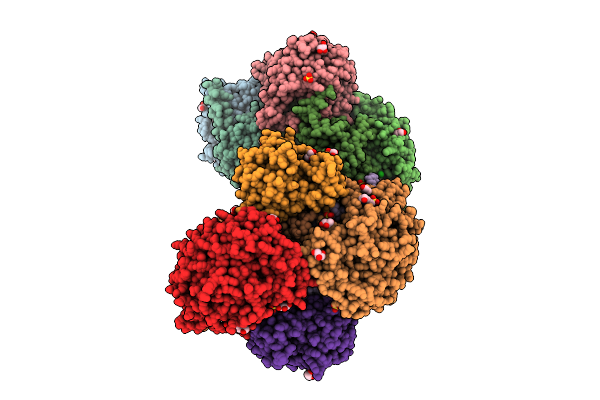
Cryo-Em Structure Of The Plpvc1 Sheath, 6-Fold Symmetrized (C6), In Contracted State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 1
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, NA, SO4, GOL, FE, SF4, FAD, CL, EPE |
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 2
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: NFU, GOL, FE, SF4, FAD |
 |
Dimeric State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus In Crystalline Form 3
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: GOL, MES, NFU, FE, SO4, EDO, FAD, SF4, PE4, FES |
 |
Cubic State Of The F420-Reducing Hydrogenase From Methanothermococcus Thermolithotrophicus
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: SO4, NFU, FAD, GOL, SF4 |
 |
A1 Tei + D-Hpg: Adenylation Domain 1 Core Construct From Teicoplanin Biosynthesis With D-4-Hydroxyphenylglycine
Organism: Actinoplanes teichomyceticus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: LIGASE Ligands: GHP, MES, BO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: LIGASE Ligands: ZN |
 |
Structure Of Rnf38 Ring With Linchpin Mutant R454Y In Complex With Ubch5B-Ub
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: LIGASE Ligands: ZN |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex Hexaguluronic Acid At Ph 3.5
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: SO4 |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex Hexamannuluronic Acid At Ph 3.5
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: SO4 |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex Pentasaccharide (Mangul)3 At Ph 3.5
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: SO4 |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex Tetraguluronic Acid At Ph 8
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: SO4 |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex Tetramannuronic Acid At Ph 8
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE |
 |
Bacteroides Ovatus Polysaccharide Lyase Family 38 (Bopl38) Wild Type In Complex With An Unsaturated Mannuronic Acid Tetramer At Ph 3.5
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: SO4 |

