Search Count: 574
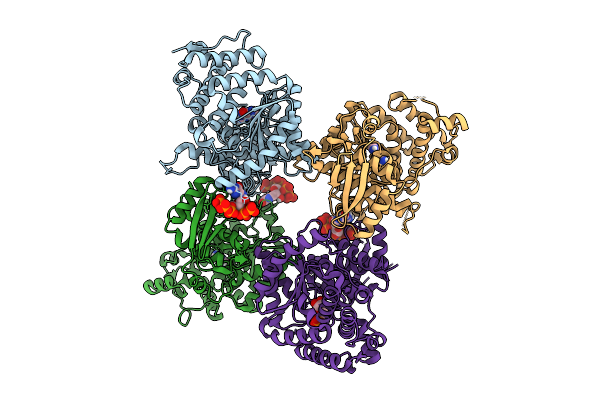 |
Crystal Structure Of The Escherichia Coli Nucleosidase Ppnn (Partial Alarmone Form)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 0O2, GUN, HSX |
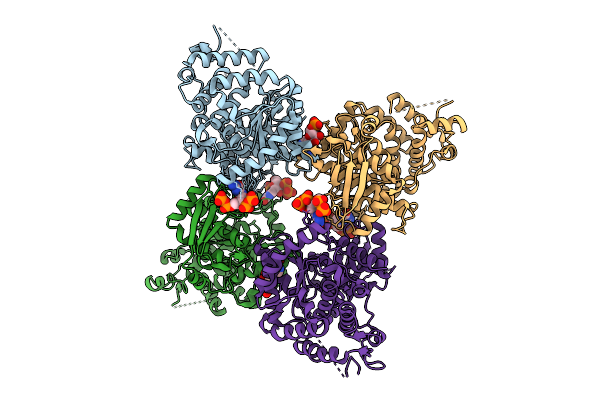 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: G4P, A1IXI |
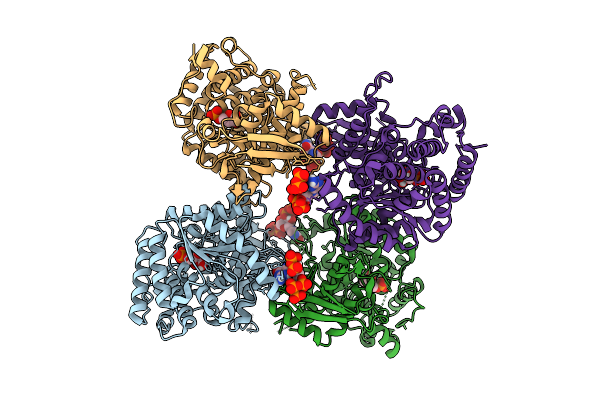 |
Crystal Structure Of The Escherichia Coli Nucleosidase Ppnn (E264A, Pppgpp Form)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: HSX, 0O2 |
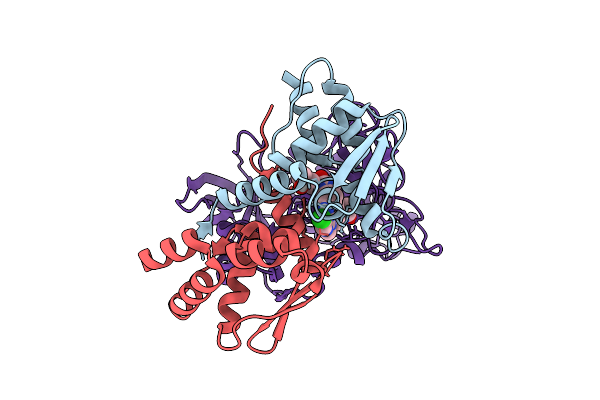 |
Cryo-Em Structure Of Ternary Complex Bcl6-Crbn-Ddb1 With Bcl6-760 (Ldd, Local Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: LIGASE Ligands: ZN, A1CM7 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH7 |
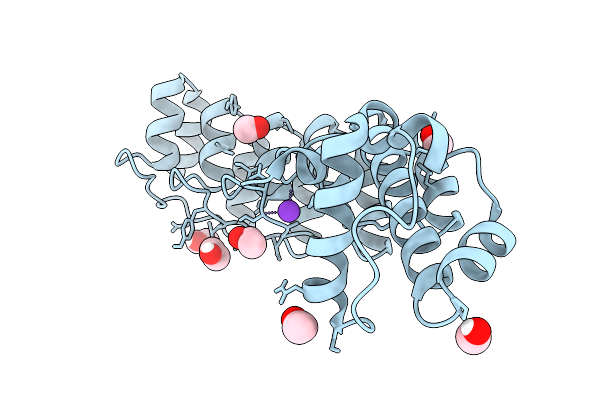 |
A Darpin Fused To The Double Trigger 1Tel Variant Crystallization Chaperone Via A Direct Helical Fusion
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, K |
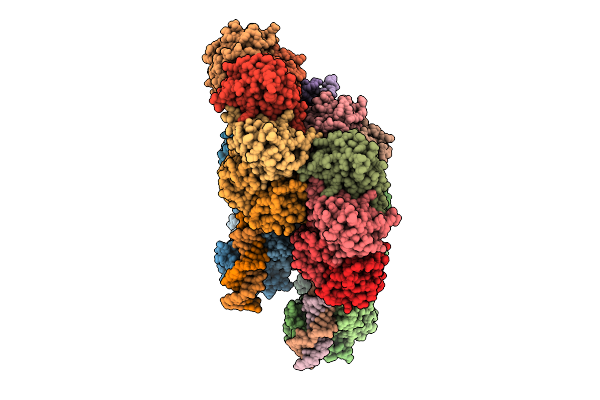 |
Crystal Structure Of The Pseudomonas Putida Xre-Res Toxin-Antitoxin Complex Bound To Promoter Dna
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: GENE REGULATION |
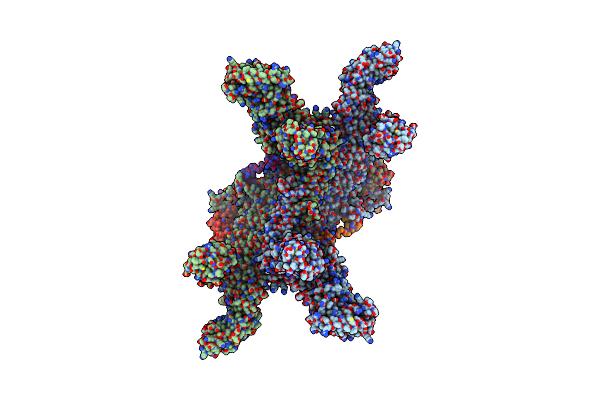 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: LIGASE Ligands: ZN, CA |
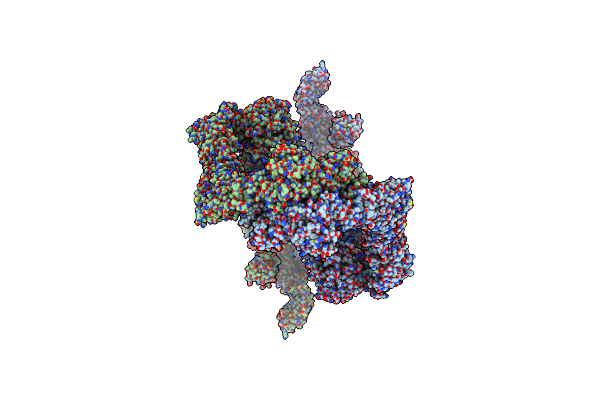 |
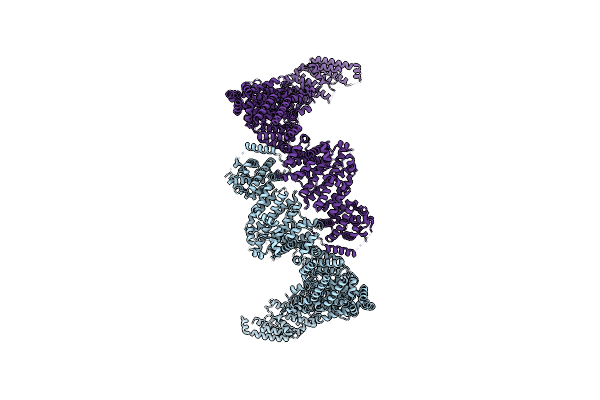
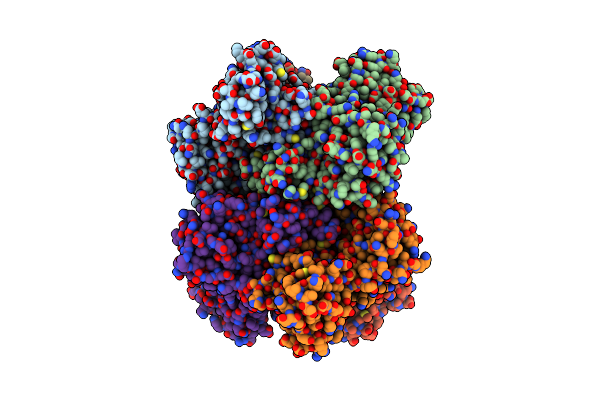
Structure Of Human Ubr4-Kcmf1-Cam E3 Ligase Complex (Silencing Factor Of The Integrated Stress Response, Sifi)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: LIGASE Ligands: ZN, CA |
 |
 |
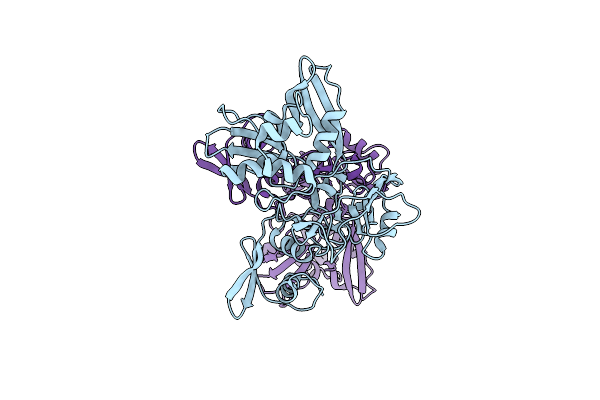
Structural Asymmetry In Sars-Cov-2 Nsp15 Hexamer Important For Catalytic Activity
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
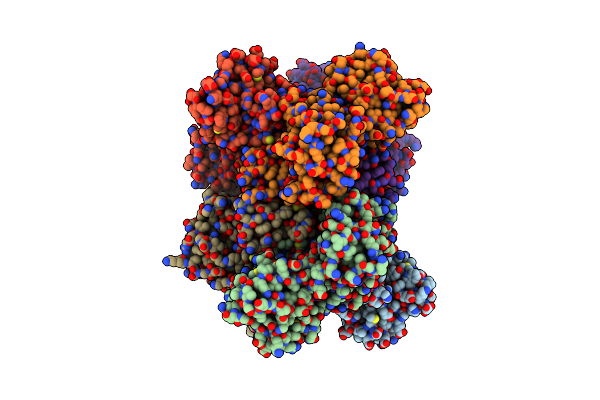
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-21 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-05-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2025-05-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2025-05-21 Classification: UNKNOWN FUNCTION |

