Search Count: 640
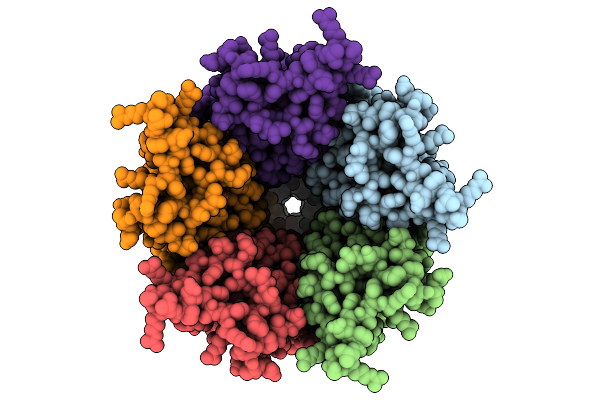 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiii Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
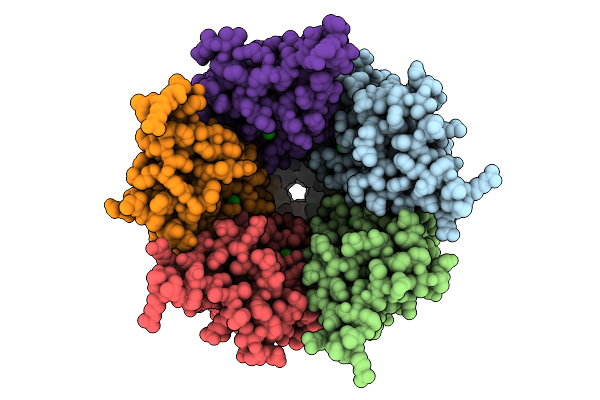 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiiio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
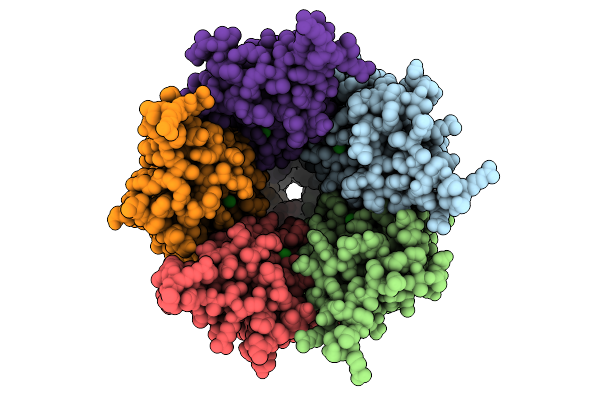 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
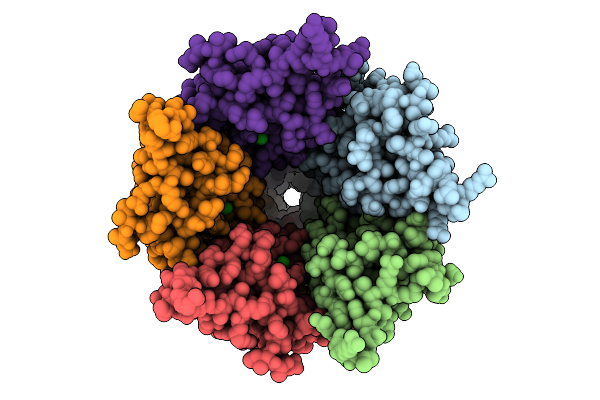 |
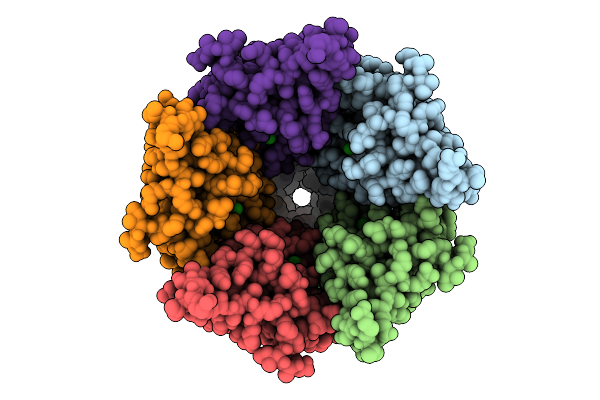
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iiooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
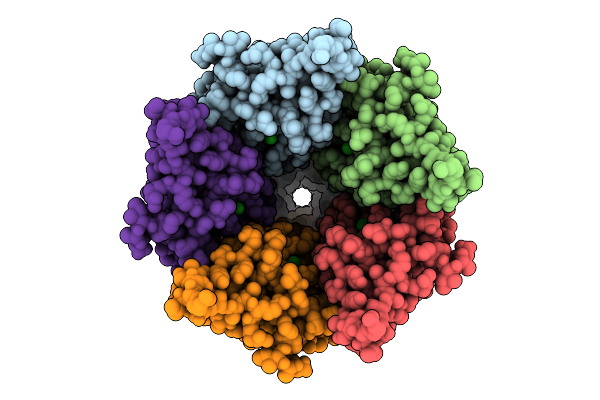
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
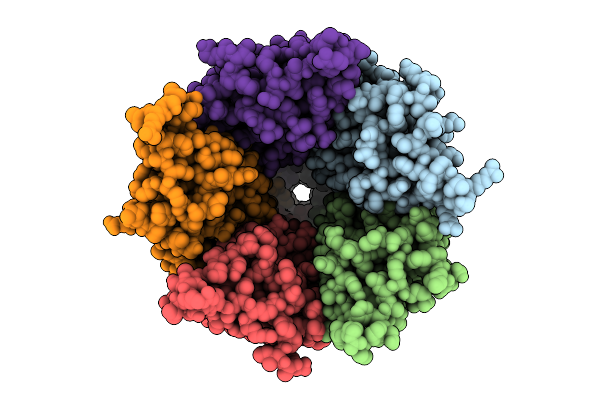
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ooooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
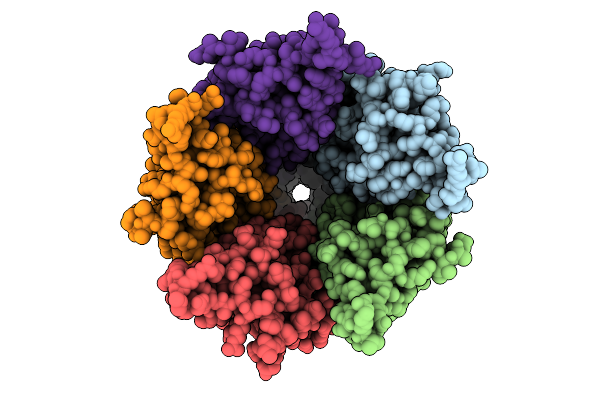
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Iioio Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Ioioo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic F116A Mutant At Ph 2.5
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic Y251A Mutant At Ph 2.5 In Ioooo Conformation
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, A1MA6 |
 |
Cryo-Em Structure Of Palytoxin-Bound Na+,K+-Atpase In The Transient State Of Dephosphorylation (E2~P)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, ALF, MG, NA, A1MA6 |
 |
Cryo-Em Structure Of Na+,K+-Atpase That Forms A Cation Channel With Palytoxin (Atp Form)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, ATP, NA, A1MA6 |
 |
Cryo-Em Structure Of Na+,K+-Atpase That Forms A Cation Channel With Palytoxin (Adp Form)
Organism: Squalus acanthias
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: CLR, PCW, MG, ADP, NA, A1MA6 |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: MG, CLR, PCW, A1MA6, NAG |
 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody.
Organism: Lama glama, Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody And Gdp-Mannose.
Organism: Candida albicans, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GDD |
 |
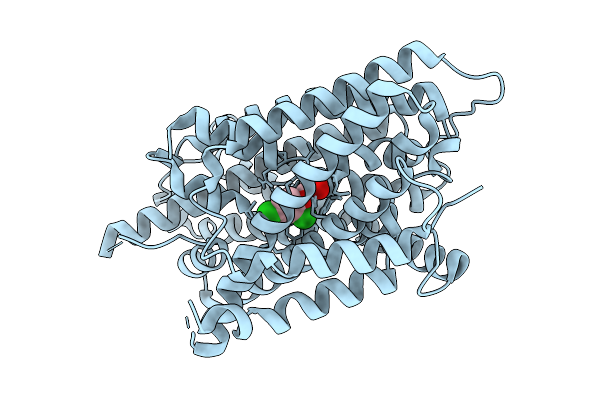
Auxin Transporter-Like Protein 3 (Lax3) In The Inward Occluded State In Complex With Auxin (Indole-3-Acetic Acid, Iaa)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: IAC |
 |
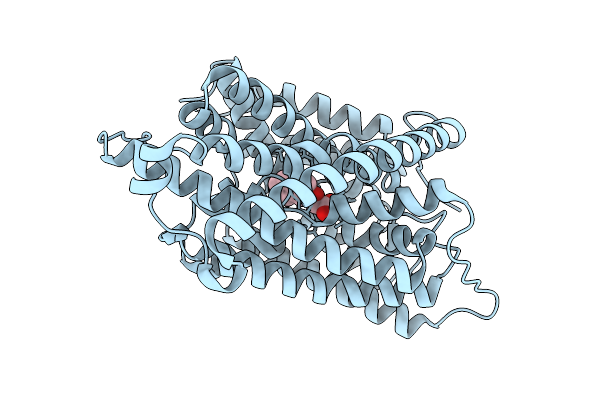
Auxin Transporter-Like Protein 3 (Lax3) In The Inward Occluded State In Complex With 2,4-Dichlorophenoxyacetic Acid (2,4-D)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: CFA |
 |
Auxin Transporter-Like Protein 3 (Lax3) In The Fully Occluded State In Complex With 2-Naphthoxyacetic Acid (2-Noa)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: A1ISN |

