Search Count: 25
 |
Crystal Structure Of An Engineered Variant Of Galactose Oxidase, Goaserd4Bb, From Fusarium Graminearum
Organism: Fusarium graminearum
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: CU, GOL |
 |
Crystal Structure Of An Engineered Variant Of Galactose Oxidase, Goaserd7Bb, From Fusarium Graminearum
Organism: Fusarium graminearum
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ACT, CU, CA |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-05-06 Classification: TRANSFERASE/ANTIBIOTIC Ligands: NMY, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-01-29 Classification: ANTIBIOTIC Ligands: CA |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-01-29 Classification: TRANSFERASE/ANTIBIOTIC Ligands: GOL, CL, MG |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-29 Classification: TRANSFERASE/ANTIBIOTIC Ligands: NMY, MG |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-01-29 Classification: TRANSFERASE/ANTIBIOTIC Ligands: NMY, APC, CA |
 |
Ternary Structure Of The E52D Mutant Of Ant-4'' With Neomycin, Amp And Pyrophosphate
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2020-01-29 Classification: TRANSFERASE/ANTIBIOTIC Ligands: NMY, PPV, MG, AMP |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-01-15 Classification: transferase/ANTIBIOTIC Ligands: NMY, MG, CA |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-15 Classification: transferase/ANTIBIOTIC Ligands: NMY, APC, MG |
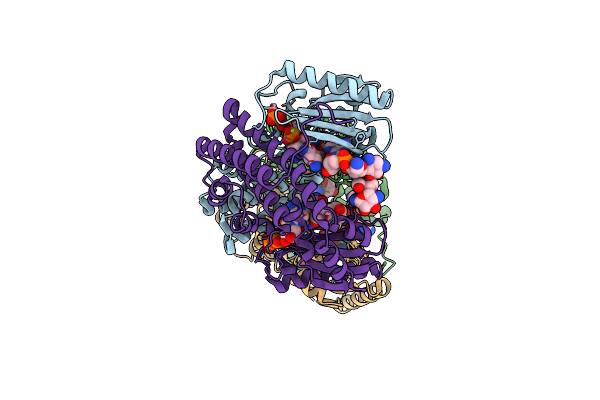 |
Ternary Complex Structure Of The T130K Mutant Of Ant-4 With Neomycin, Ampcpp And Pyrophosphate
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-01-15 Classification: transferase/ANTIBIOTIC Ligands: NMY, AMP, PPV, MG |
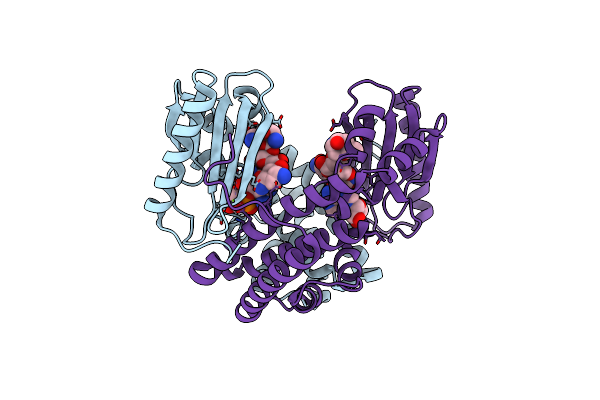 |
Ternary Complex Structure Of The T130K Mutant Of Ant-4'' With Neomycin And Atp (No Pyrophosphate)
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-01-15 Classification: transferase/ANTIBIOTIC Ligands: NMY, AMP, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2019-12-18 Classification: DE NOVO PROTEIN Ligands: 308, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.923 Å, 2.500 Å Release Date: 2019-12-18 Classification: DE NOVO PROTEIN Ligands: 308, NA, DOD |
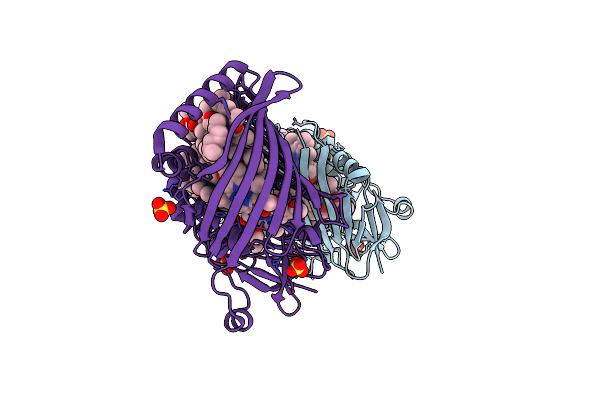 |
X-Ray Structure Of The Fenna-Matthews-Olsen Antenna Complex From Prosthecochloris Aestuarii
Organism: Prosthecochloris aestuarii
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2019-03-13 Classification: PHOTOSYNTHESIS Ligands: BCL, SO4 |
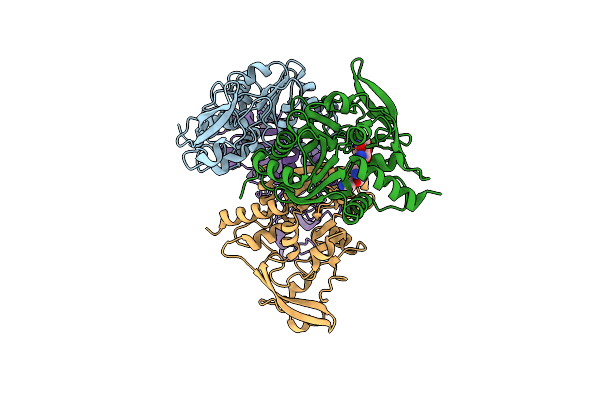 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: SIS |
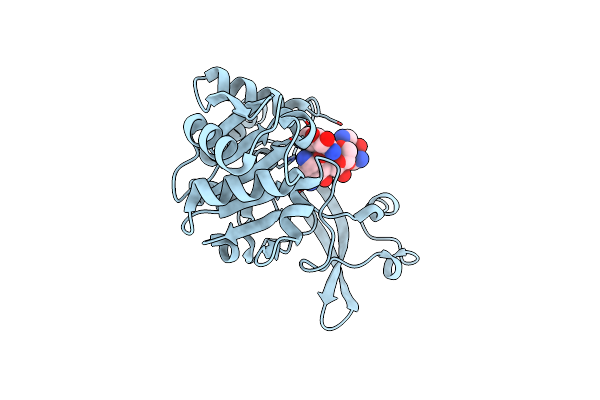 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: NMY |
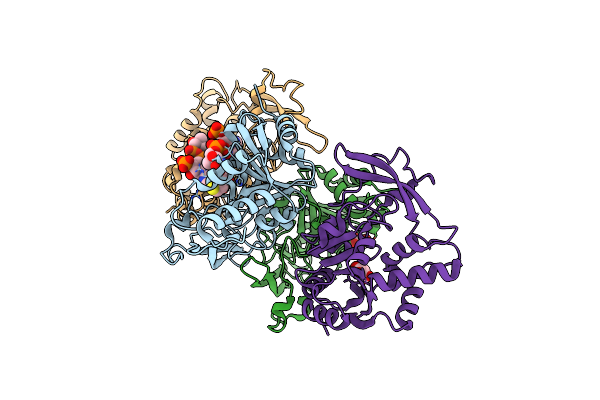 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-11-07 Classification: TRANSFERASE Ligands: COA, MLI |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: PAR |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-11-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: CL |

