Search Count: 70
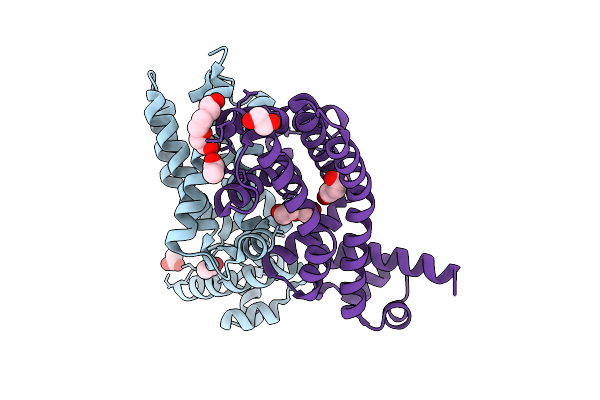 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2025-11-26 Classification: TRANSCRIPTION Ligands: ACT, PEG, CL, PGE, EDO |
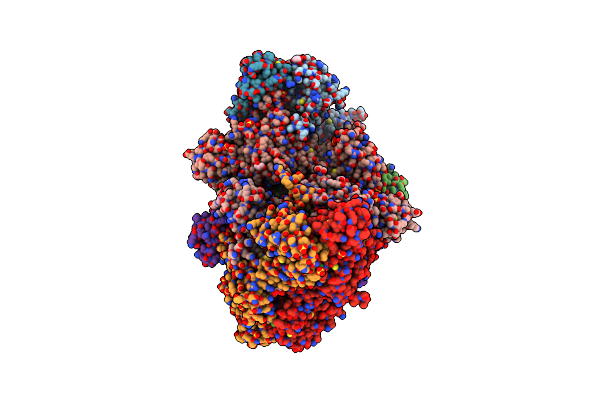 |
Crystal Structure Of Argininosuccinate Lyase From Arabidopsis Thaliana (Atasl)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-10-23 Classification: LYASE Ligands: SO4, GOL, CL, NA |
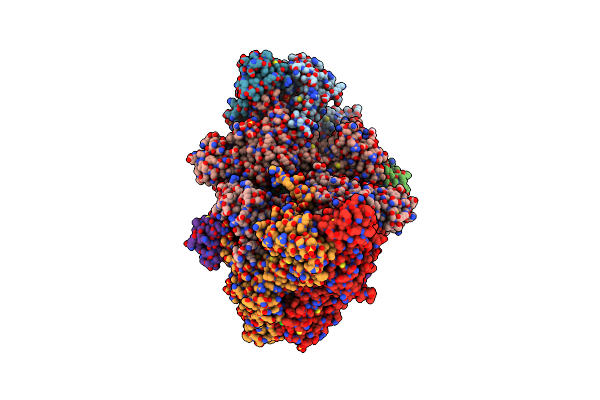 |
Crystal Structure Of Argininosuccinate Lyase From Arabidopsis Thaliana (Atasl) In Complex With Biological Substrate And Products - Argininosuccinate, Argnine And Fumarate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-23 Classification: LYASE Ligands: AS1, SO4, CL, FUM, ARG, GOL |
 |
Crystal Structure Of Ornithine Transcarbamylase From Arabidopsis Thaliana (Atotc) In Complex With Ornithine
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: ORN, PEG, EDO, SO4, NA |
 |
Crystal Structure Of Ornithine Transcarbamylase From Arabidopsis Thaliana (Atotc) In Complex With Carbamoyl Phosphate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: CP, EDO, PGE, PEG, SO4, NA |
 |
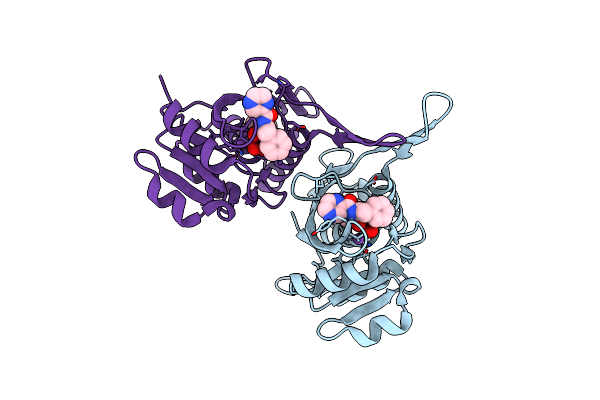
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-05-17 Classification: TRANSPORT PROTEIN Ligands: 8XI, MLI, LMR, FMT, ACT |
 |
Crystal Structure Of Atp-Dependent Lon Protease From Bacillus Subtillis (Bslonba)
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: BO2, NA |
 |
Crystal Structure Of Serine Hydroxymethyltransferase, Isoform 2 From Arabidopsis Thaliana (Shm2)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: EDO, CL, EPE, PEG, SO4 |
 |
Crystal Structure Of Serine Hydroxymethyltransferase, Isoform 4 From Arabidopsis Thaliana (Shm4)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: EDO, TRS |
 |
Crystal Structure Of Serine Hydroxymethyltransferase, Isoform 6 From Arabidopsis Thaliana (Shm6)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-08-24 Classification: TRANSFERASE Ligands: NO3 |
 |
Crystal Structure Of Serine Hydroxymethyltransferase, Isoform 7 From Arabidopsis Thaliana (Shm7)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-08-24 Classification: TRANSFERASE |
 |
Crystal Structure Of Catalase Hpii From Escherichia Coli (Serendipitously Crystallized)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: HDD, GOL, EDO |
 |
Crystal Structure Of Catalase Hpii From Escherichia Coli (Serendipitously Crystallized)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: HDD, TRS, GOL, MPD, EDO |
 |
Crystal Structure Of Catalase Hpii From Escherichia Coli (Serendipitously Crystallized)
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: GOL, EDO, HDD |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-09-02 Classification: RNA Ligands: MG, K |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-09-02 Classification: RNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2020-09-02 Classification: RNA |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: MN |
 |
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: MN, ORN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: MN, NA, ORN, GOL, PEG |

