Search Count: 33
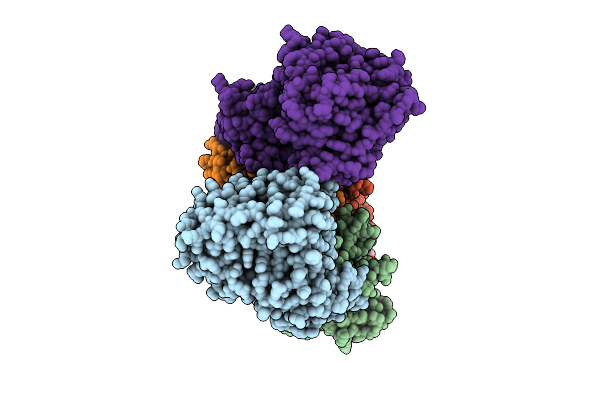 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Middle State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, LMT, FES, FAD |
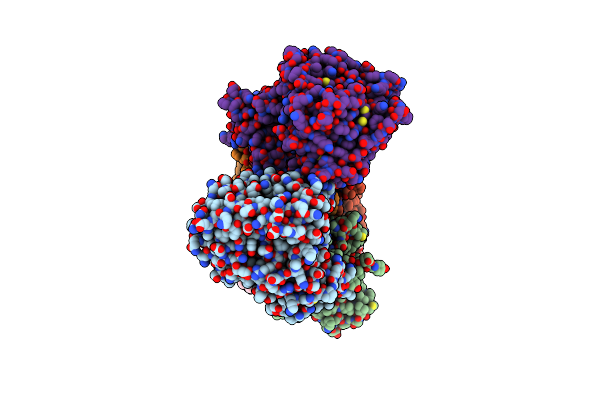 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
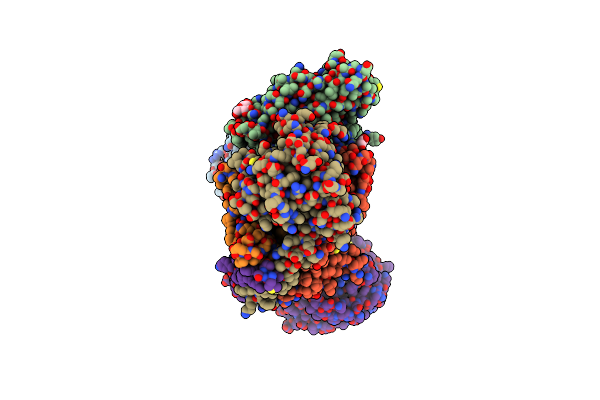 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
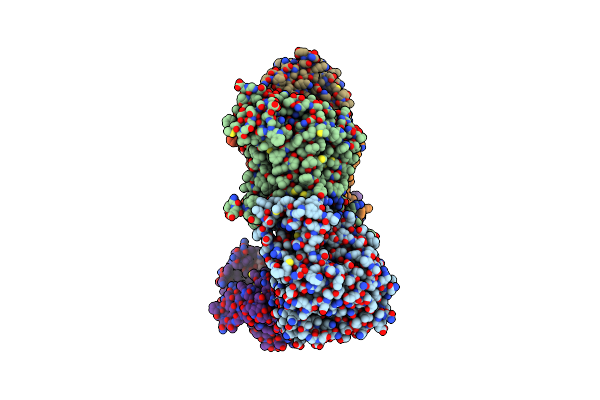 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, FMN, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, CA, FMN, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Upper State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Down State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Stable State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Shifted State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, PEE, LMT, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Aurachin D-42
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, 0NI, PEE, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD, NAI |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: HYDROLASE Ligands: PGE, NAG, K |
 |
Acetyl Xylan Esterase B From Aspergillus Oryzae (Aoaxeb), Succinate Complex Form
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: HYDROLASE Ligands: NAG, ACT, SIN |
 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: HYDROLASE Ligands: NAG |
 |
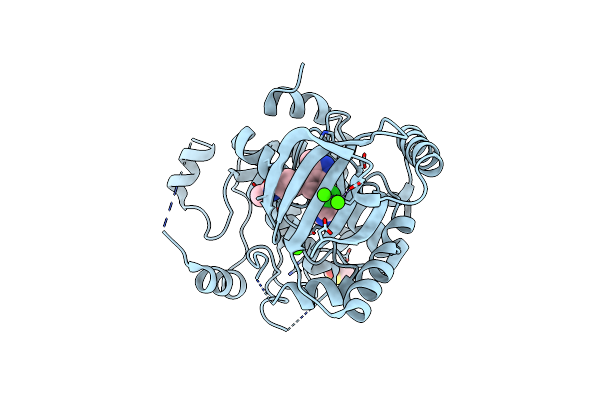
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: FZC, DMS, CA |
 |
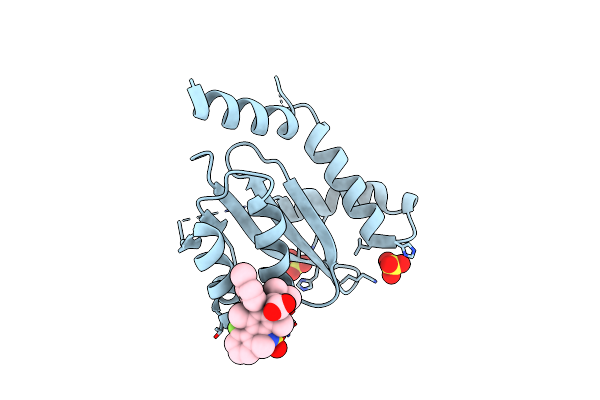
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-01 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GIK, DMS, CA |
 |
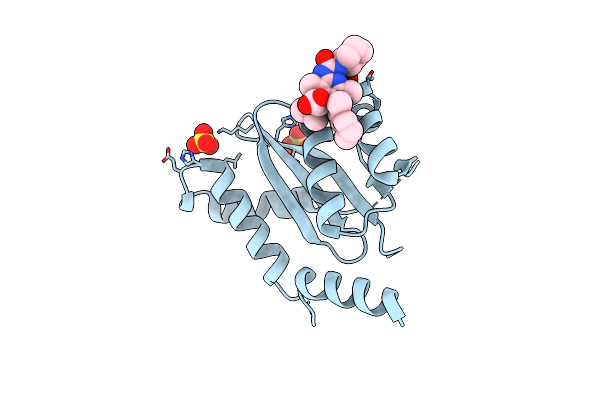
Crystal Structure Of Hiv-1 Integrase Catalytic Core Domain In Complex With (2S)-2-(Tert-Butoxy)-2-(10-Fluoro-2-(2-Hydroxy-4-Methylphenyl)-1,4-Dimethyl-5-(Methylsulfonyl)-5,6-Dihydrophenanthridin-3-Yl)Acetic Acid
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: SO4, 8Z3, GOL |
 |
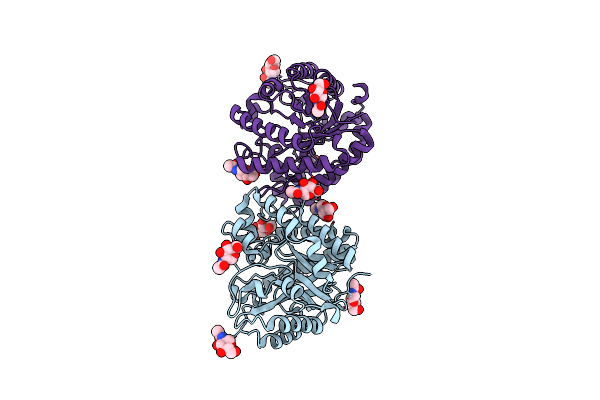
Crystal Structure Of Hiv-1 Integrase Catalytic Core Domain In Complex With 2-(Tert-Butoxy)-2-(2-(3-Cyclohexylureido)-3,6-Dimethyl-5-(5-Methylchroman-6-Yl)Pyridin-4-Yl)Acetic Acid
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate ny5)
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-01-13 Classification: VIRAL PROTEIN Ligands: SO4, GZ9 |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: NAG |

