Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
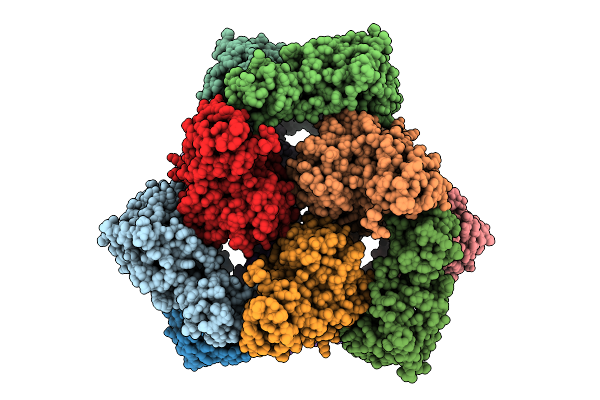 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSPORT PROTEIN |
 |
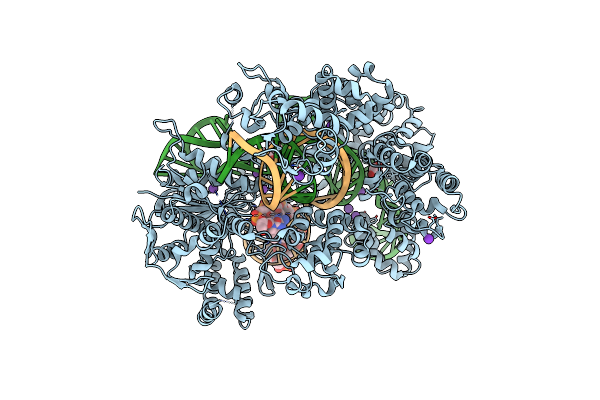
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: NO3 |
 |
Organism: Streptococcus pyogenes serotype m1, Streptococcus pyogenes
Method: X-RAY DIFFRACTION Release Date: 2018-10-31 Classification: HYDROLASE/RNA/DNA Ligands: K, MG, EDO |
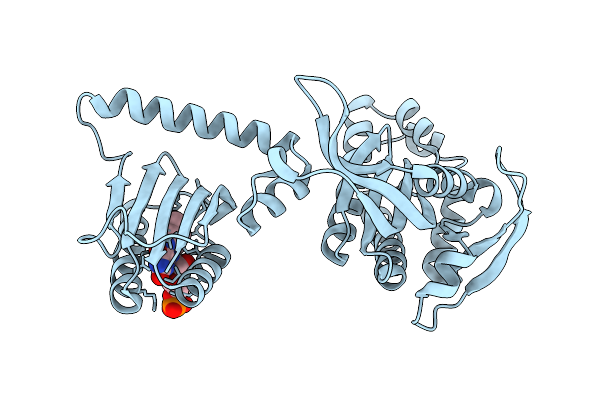 |
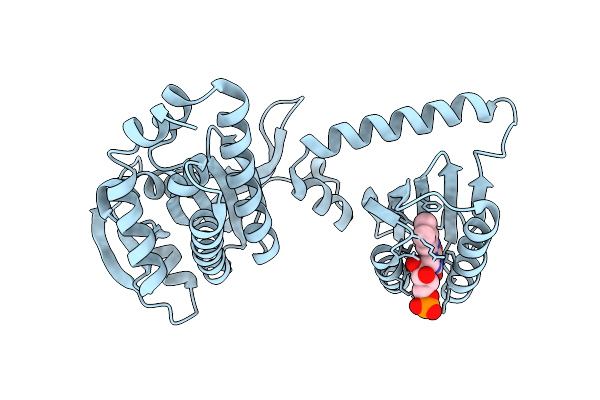
Organism: Cyanobacteria
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-07-26 Classification: LYASE Ligands: FMN |
 |
Organism: Cyanobacteria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-26 Classification: LYASE Ligands: FMN |
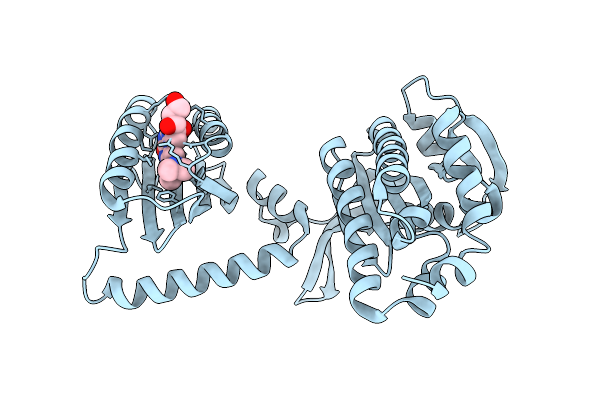 |
Organism: Cyanobacteria
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-26 Classification: LYASE Ligands: RS3 |
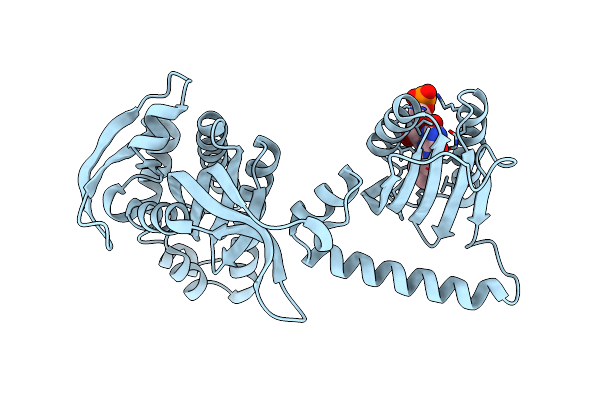 |
Crystal Structure Of Photoactivated Adenylyl Cyclase Of A Cyanobacteriaoscillatoria Acuminata In Hexagonal Form
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-01 Classification: LYASE Ligands: FMN |
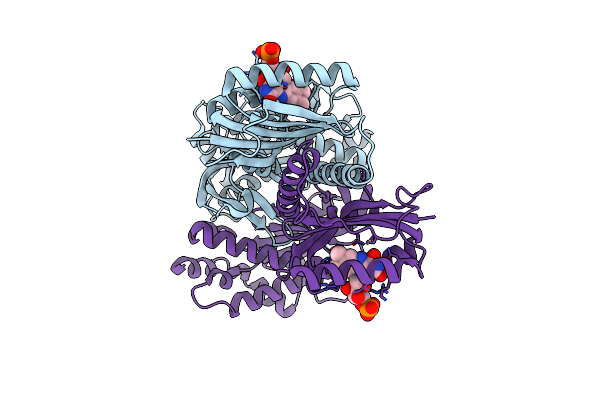 |
Crystal Structure Of Photoactivated Adenylyl Cyclase Of A Cyanobacteriaoscillatoria Acuminata In Orthorhombic Form
Organism: Oscillatoria acuminata pcc 6304
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-06-01 Classification: LYASE Ligands: FMN |
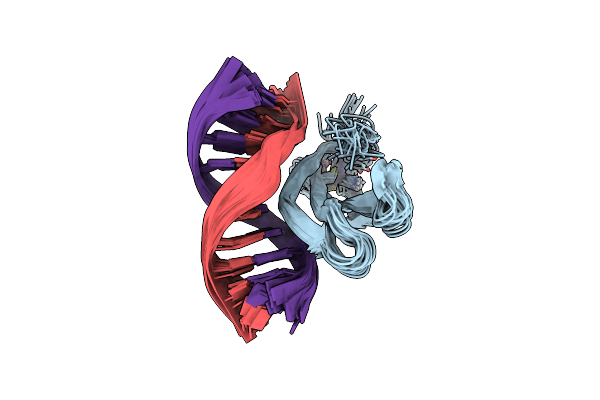 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2012-01-18 Classification: TRANSCRIPTION/DNA Ligands: ZN |
 |
Nmr Structure Of The Hypothetical Enth-Vhs Domain At3G16270 From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2005-05-03 Classification: structural genomics, unknown function |
 |
Nmr Structure Of The Hypothetical Rhodanese Domain At4G01050 From Arabidopsis Thaliana
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2005-01-25 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: DNA BINDING PROTEIN |
 |
Solution Structure Of The Dna-Binding Domain Of Ethylene-Insensitive3-Like3
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: DNA BINDING PROTEIN |
 |
Solution Structure Of The Dna-Binding Domain Of Squamosa Promoter Binding Protein-Like 12 Lacking The Second Zinc-Binding Site
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-11-28 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Solution Structure Of The Dna-Binding Domain Of Squamosa Promoter Binding Protein-Like 4
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-03-09 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Solution Structure Of The Dna-Binding Domain Of Squamosa Promoter Binding Protein-Like 7
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-03-09 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Solution Structure Of The Fha Domain Of Arabidopsis Thaliana Hypothetical Protein
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2004-01-10 Classification: structural genomics, unknown function |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2003-12-16 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |