Search Count: 62
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanogenic Archaeon Iso4-G1
Organism: Methanogenic archaeon mixed culture iso4-g1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2023-03-15 Classification: TRANSLATION |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-22 Classification: TRANSLATION,LIGASE |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Mazzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TR, K, PEG, PGE |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Mtmdzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TU, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Metzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9TX, K, PEG |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Tco*Lys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U0, K, PEG |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Zaesecys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U6 |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Bcnlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9U9, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Pno2Zlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9UC, CIT, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Pampylys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9UF, K, PO4 |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Oazzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9V0, K |
 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Oclzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9V6, K, EDO |
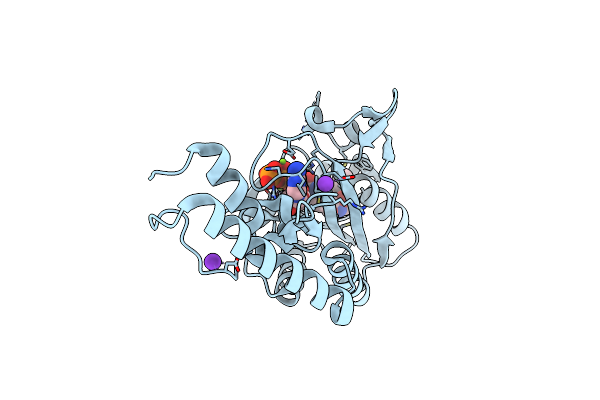 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Zlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9VC, K |
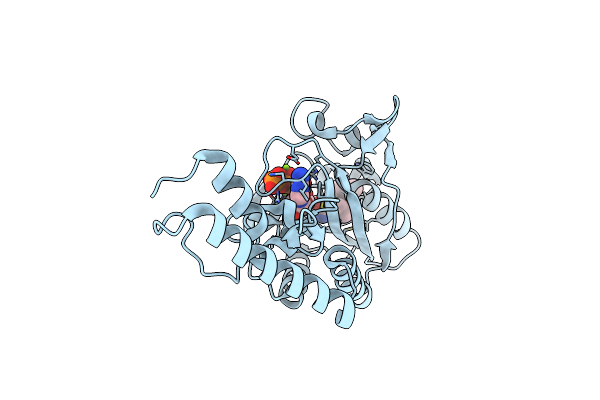 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Teoclys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9VF |
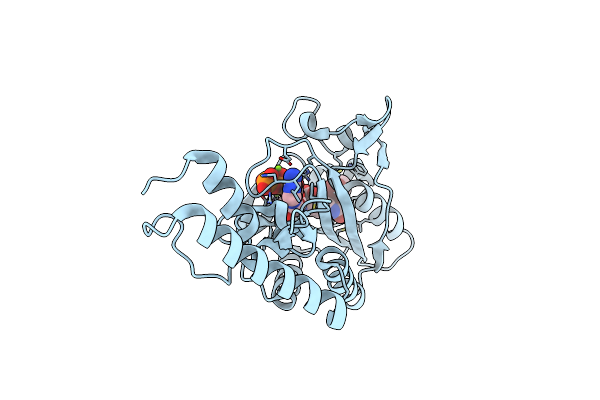 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Obrzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9VL |
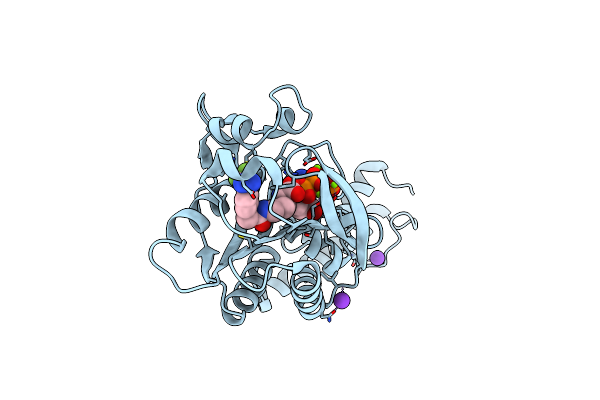 |
Crystal Structure Of Methanosarcina Mazei Pylrs(Y306A/Y384F) Complexed With Ptmdzlys
Organism: Methanosarcina mazei jcm 9314
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-04-17 Classification: TRANSLATION Ligands: MG, ATP, 9VR, K |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-09-15 Classification: Metal-binding protein/nuclear Protein Ligands: ZN |
 |
Solution Structure Of Tandem Zinc Finger Domain 12 In Muscleblind-Like Protein 2
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2009-05-12 Classification: TRANSCRIPTION Ligands: ZN |
 |
Solution Structure Of The Zf-Ccchx2 Domain Of Muscleblind-Like 2, Isoform 1 [Homo Sapiens]
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2007-06-26 Classification: TRANSCRIPTION Ligands: ZN |
 |
Solution Structure Of The Zf-Cw Domain In Zinc Finger Cw-Type Pwwp Domain Protein 1
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2007-06-26 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN |

