Search Count: 87
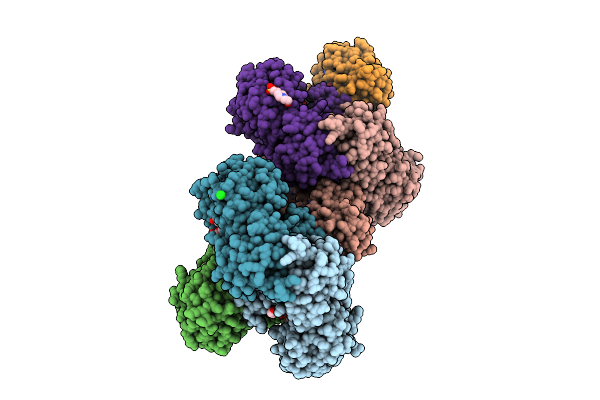 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad And Glyceraldehyde-3-Phosphate
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: OXIDOREDUCTASE Ligands: G3H, PEG, NAD, NA, PGE, CL, GOL, EPE |
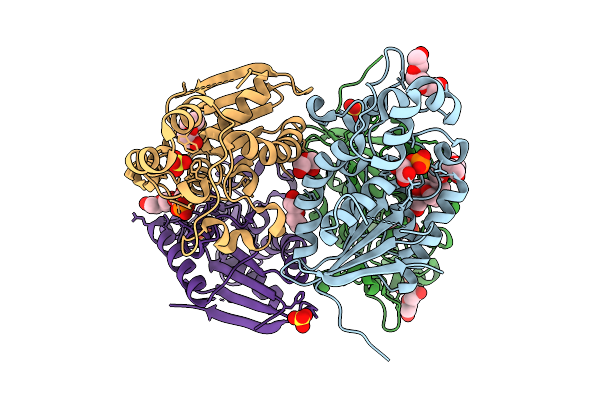 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis In Complex With 3-Phosphoglyceric Acid
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ISOMERASE Ligands: CL, 3PG, PG4, SO4 |
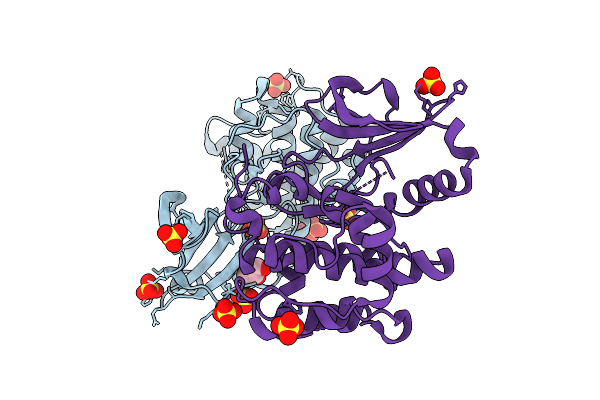 |
Crystal Structure Of Serine/Threonine-Protein Kinase (Aek1) From Trypanosoma Brucei
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SO4, CL, GOL |
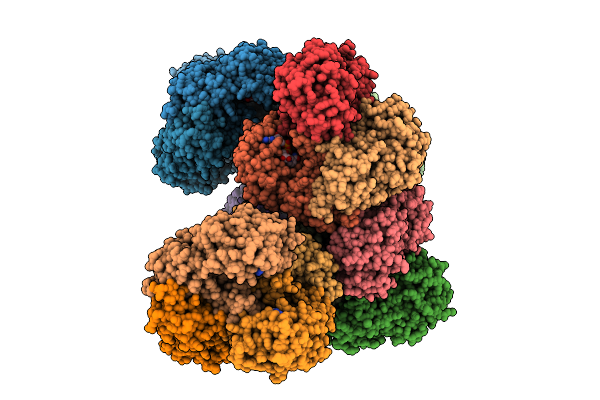 |
Crystal Structure Of A Glyceraldehyde-3-Phosphate Dehydrogenase From Neisseria Gonorrhoeae In Complex With Nad (P1 Form)
Organism: Neisseria gonorrhoeae nccp11945
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: NAD, PGE |
 |
Crystal Structure Of 6,7-Dimethyl-8-Ribityllumazine Synthase From Bordetella Pertussis In Complex With 5-Amino-6-(D-Ribitylamino)Uracil
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: CL, LMZ |
 |
Crystal Structure Of Prolyl-Trna Synthetase (Prors, Proline--Trna Ligase) From Plasmodium Falciparum In Complex With Inhibitor Ynw69
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: CL, SO4, A1CYM |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Utp
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: MG, UTP |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Uridine-5'-Diphosphate-Glucose
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: SO4, MG, UPG, GOL |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis In Complex With Uridine-5'-Diphosphate-Glucose (Twinned Lattice)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: UPG, MG |
 |
Crystal Structure Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis (Sulfate Bound)
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: SO4, CL, GOL |
 |
Crystal Structure Of Capsular Polysaccharide Biosynthesis Protein From Bordetella Pertussis In Complex With Nad And Uridine-Diphosphate-N-Acetylgalactosamine
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ISOMERASE Ligands: CL, NAD, UD2, ACT |
 |
Crystal Structure Of Gtp Cyclohydrolase 1 (Fole) From Mycobacterium Tuberculosis
Organism: Plasmodium vivax sal-1
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: ZN, CL |
 |
Crystal Structure Of Structure Of Wt Bfrb From Pseudomonas Aeruginosa In Complex With A Protein-Protein Interaction Inhibitor Km-5-25
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: K, HEM, PG4, A1BYB |
 |
Crystal Structure Of Structure Of Wt Bfrb From Pseudomonas Aeruginosa In Complex With A Protein-Protein Interaction Inhibitor Km-5-35
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: HEM, A1BYC, K, NA, PG4 |
 |
Crystal Structure Of Utp--Glucose-1-Phosphate Uridylyltransferase From Bordetella Pertussis
Organism: Bordetella pertussis tohama i
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-04-30 Classification: TRANSFERASE Ligands: GLY, GOL |
 |
Crystal Structure Of Tryptophanyl-Trna Synthetase From Klebsiella Aerogenes
Organism: Klebsiella aerogenes kctc 2190
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-23 Classification: LIGASE Ligands: IMD, CA |
 |
Crystal Structure Of Tryptophanyl-Trna Synthetase From Klebsiella Aerogenes (Tryptophan Bound)
Organism: Klebsiella aerogenes kctc 2190
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-23 Classification: LIGASE Ligands: TRP, CL, SO4, PG4 |
 |
Crystal Structure Of Phosphoglycerate Mutase From Trichomonas Vaginalis (Sulfate Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-04-23 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Klebsiella aerogenes kctc 2190
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: CL |
 |
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-04-02 Classification: ISOMERASE Ligands: SO4, GOL, P33, PG4 |

