Search Count: 35
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2018-10-10 Classification: HYDROLASE Ligands: CL, NA, ACT, EDO |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-10-10 Classification: ANTIBIOTIC Ligands: NXL |
 |
Serial Time Resolved Crystallography Of Photosystem Ii Using A Femtosecond X-Ray Laser. The S State After Two Flashes (S3)
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: OEX, FE2, CLA, PHO, BCR, SQD, CL, BCT, PL9, LMG, CA, DGD, LHG, HEM, MG |
 |
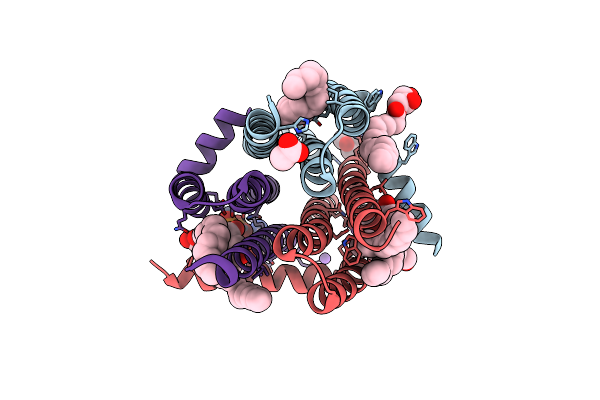
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: OLC, MPD, NO3, NA |
 |
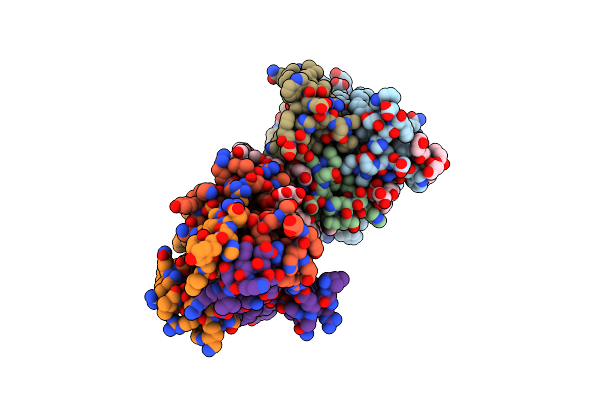
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: CL, NA, ACT, OLC, MPD, ZN, ACP |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: 79N, 79M, ZN, FLC, ACT |
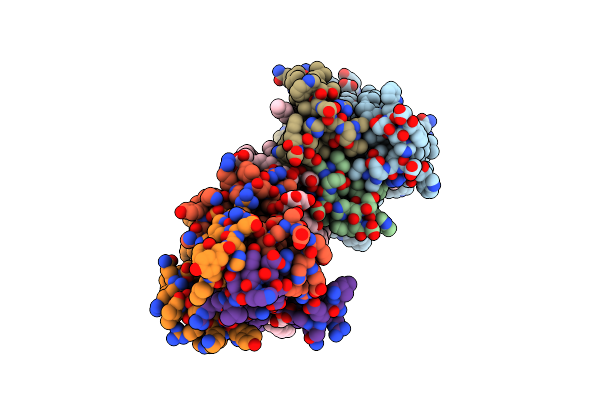 |
Structure Of Delta7-Dgka In 7.9 Mag By Serial Femtosecond Crystatallography To 2.18 Angstrom Resolution
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-09-30 Classification: TRANSFERASE Ligands: 79N, 79M, ZN, FLC |
 |
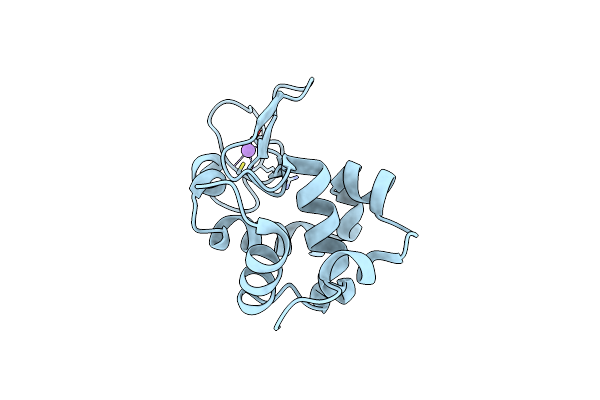
Crystal Structure Of The Sr Ca2+-Atpase In The Ca2-E1-Mgamppcp Form Determined By Serial Femtosecond Crystallography Using An X-Ray Free-Electron Laser.
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: CA, K, ACP |
 |
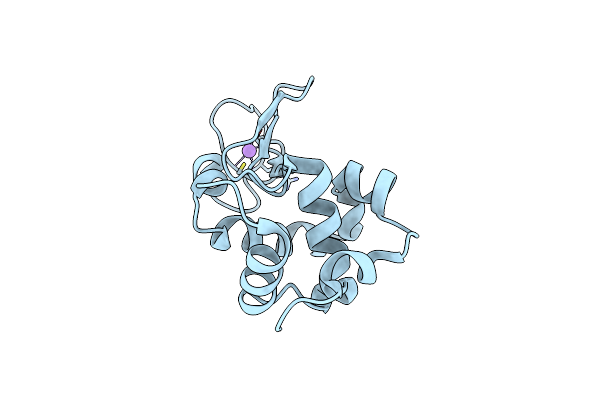
Hen Egg-White Lysozyme Structure From A Spent-Beam Experiment At Lcls: Original Beam
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-20 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure From A Spent-Beam Experiment At Lcls: Refocused Beam
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-05-20 Classification: HYDROLASE |
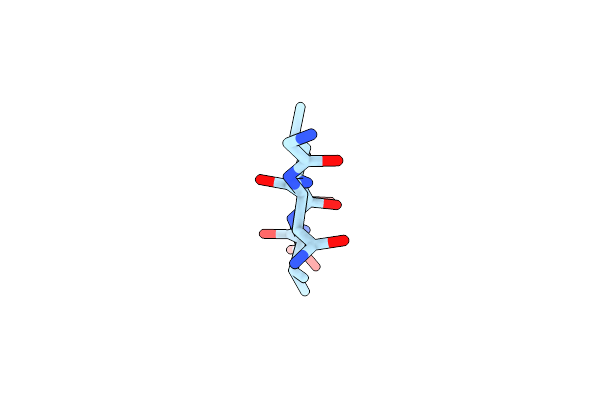 |
Structure Of The Amyloid Forming Peptide Gnlvs (Residues 26-30) From The Eosinophil Major Basic Protein (Embp)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-03-18 Classification: PROTEIN FIBRIL |
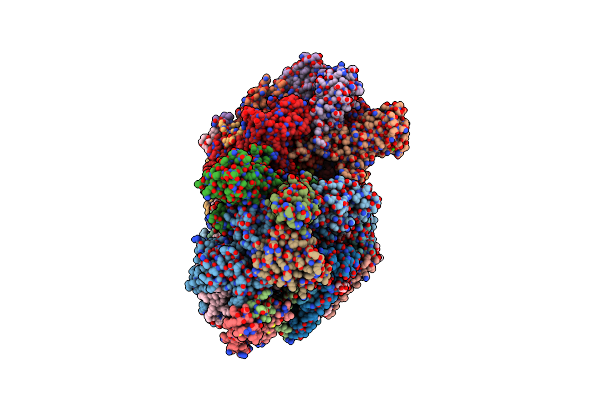 |
Serial Time-Resolved Crystallography Of Photosystem Ii Using A Femtosecond X-Ray Laser The S1 State
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:5.00 Å Release Date: 2014-07-16 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, CL, BCT, CLA, PHO, BCR, PL9, SQD, LHG, CA, DGD, HEM, MG |
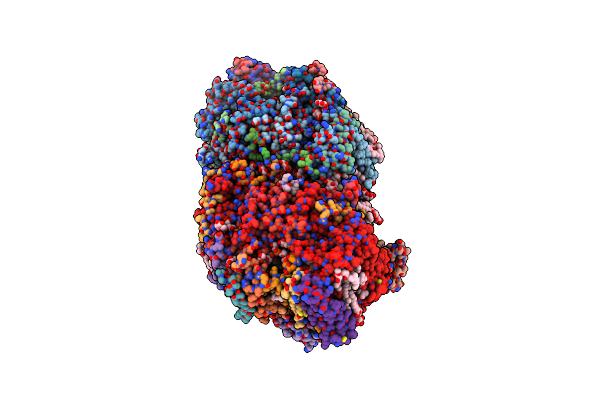 |
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.90 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, CL, OEX, SQD, LMT, PHO, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 500 Ms After The Third Illumination At 4.6 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.60 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, OEX, SQD, LMT, PHO, CL, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 500 Ms After The 2Nd Illumination (2F) At 4.5 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PL9, BCR, DGD, LHG, LMG, CL, OEX, SQD, LMT, PHO, BCT, HEM, CA |
 |
Rt Xfel Structure Of Photosystem Ii 250 Microsec After The Third Illumination At 5.2 A Resolution
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:5.20 Å Release Date: 2014-07-09 Classification: ELECTRON TRANSPORT,PHOTOSYNTHESIS Ligands: FE2, CLA, PHO, PL9, BCR, DGD, LHG, CL, OEX, SQD, LMG, LMT, BCT, HEM, CA |
 |
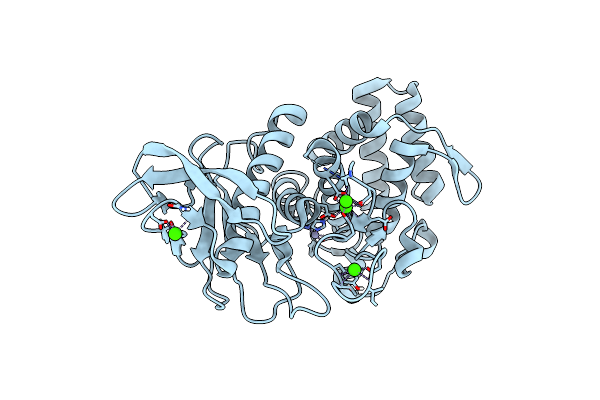
1.8 A Resolution Room Temperature Structure Of Thermolysin Recorded Using An Xfel
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: ZN, CA |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-12 Classification: HYDROLASE Ligands: ZN, CA |
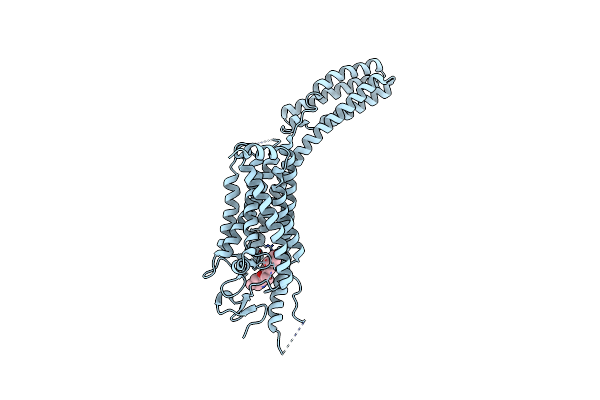 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-03-05 Classification: SIGNALING PROTEIN Ligands: CY8 |
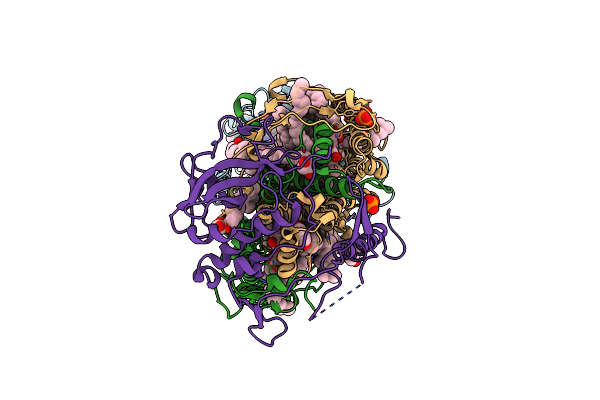 |
Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Center
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Release Date: 2013-12-25 Classification: PHOTOSYNTHESIS Ligands: HEC, DGA, BCL, BPB, MPG, FE2, MQ7, NS5, OTP, PO4 |

