Search Count: 6
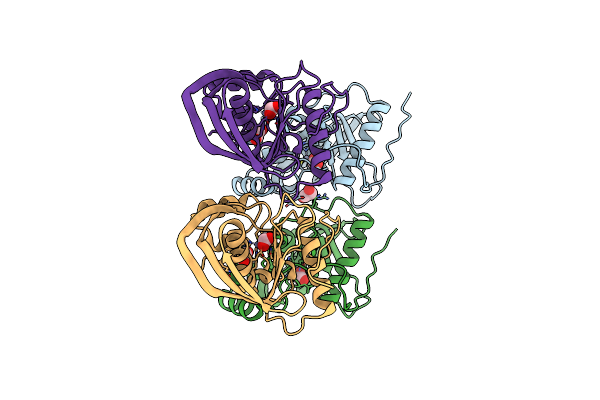 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-03-06 Classification: HYDROLASE Ligands: ZN, EDO, FMT, CL |
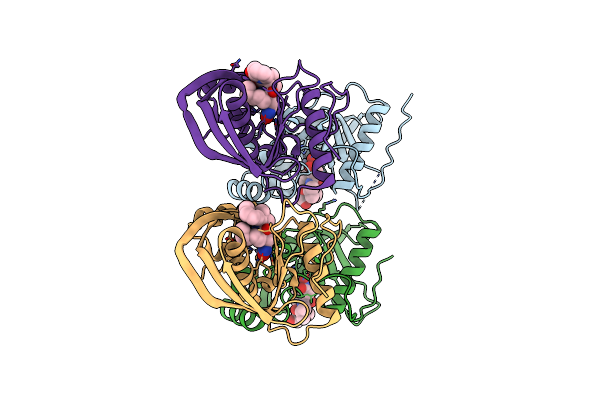 |
Crystal Structure Of A Peptide Deformylase From Synechococcus Elongatus In Complex With Actinonin
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, BR, BB2 |
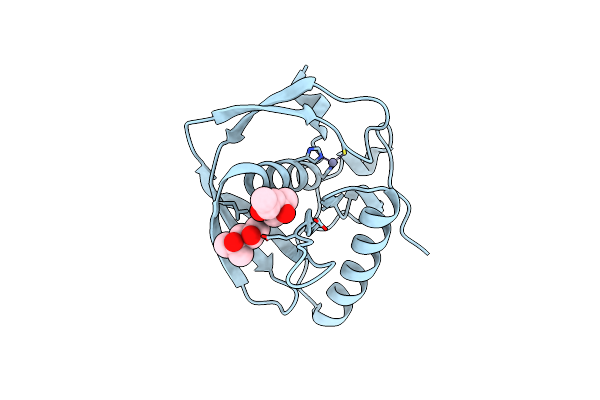 |
Crystal Structure Of A Probable Peptide Deformylase From Synechococcus Phage S-Ssm7
Organism: Synechococcus phage s-ssm7
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: ZN, MRD |
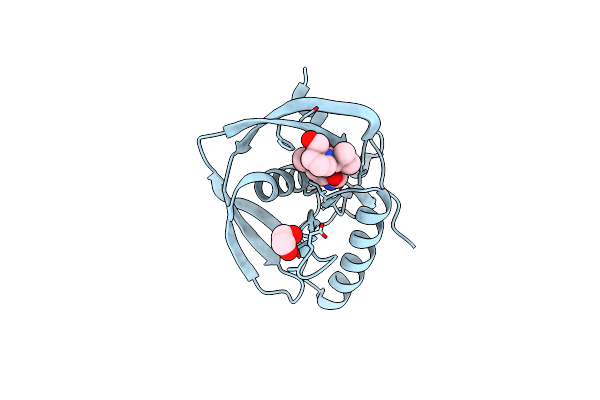 |
Crystal Structure Of A Probable Peptide Deformylase From Strucynechococcus Phage S-Ssm7 In Complex With Actinonin
Organism: Synechococcus phage s-ssm7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-01-09 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, CL, EDO, BB2 |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-12-14 Classification: Hydrolase, Ligase/DNA |
 |
High Resolution Structure Of The Holliday Junction Intermediate In Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-12-14 Classification: HYDROLASE, LIGASE/DNA |

