Search Count: 35
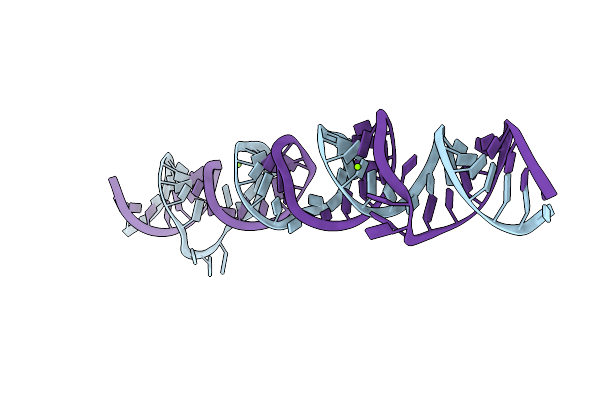 |
Dimerized Structure Gives Further Insight Into The Function Of The Novel Rna Gene: Har1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-03-29 Classification: RNA Ligands: MG |
 |
Structural Variations And Solvent Structure Of Uggggu Quadruplexes Stabilized By Sr2+ Ions
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.88 Å Release Date: 2014-11-19 Classification: RNA Ligands: SR, NA, CA |
 |
Structural Variations And Solvent Structure Of Uggggu Quadruplexes Stabilized By Sr2+ Ions
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2014-11-19 Classification: RNA Ligands: SR, NA, CA |
 |
Structural Variations And Solvent Structure Of Uggggu Quadruplexes Stabilized By Sr2+ Ions
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2014-11-12 Classification: RNA Ligands: SR, NA, CA |
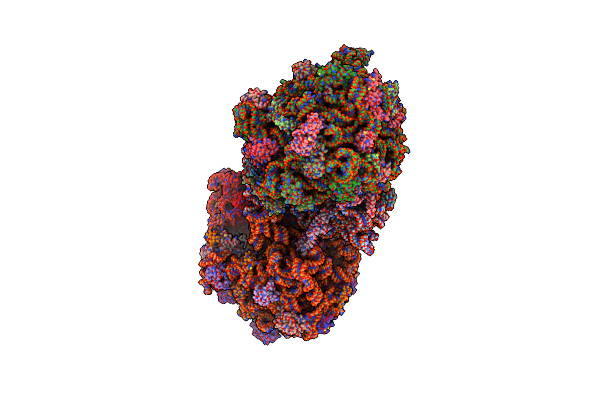 |
Crystal Structure Of A Translation Termination Complex Formed With Release Factor Rf2.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-03-06 Classification: RNA |
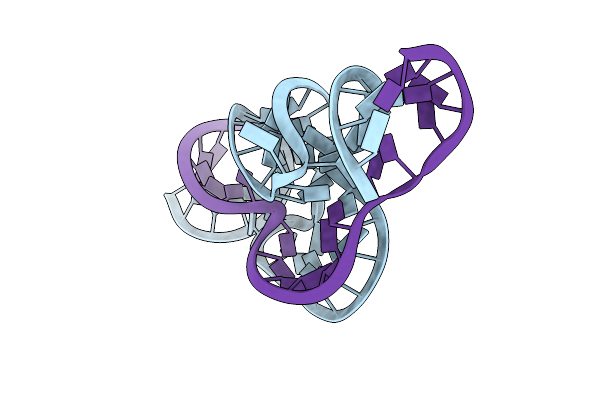 |
Full-Length Hammerhead Ribozyme With G12A Substitution At The General Base Position
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-12-12 Classification: RNA |
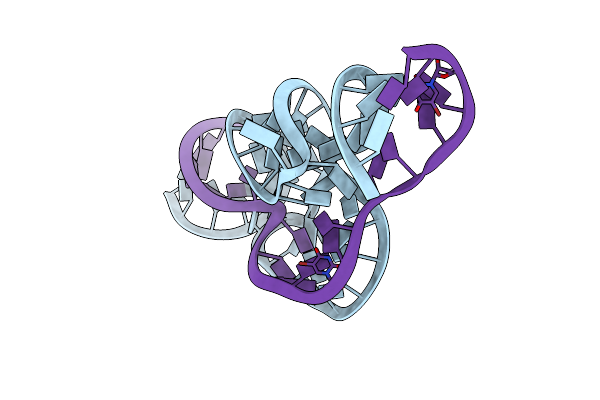 |
The 2.2 A Structure Of A Full-Length Catalytically Active Hammerhead Ribozyme
Organism: schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-12-12 Classification: RNA |
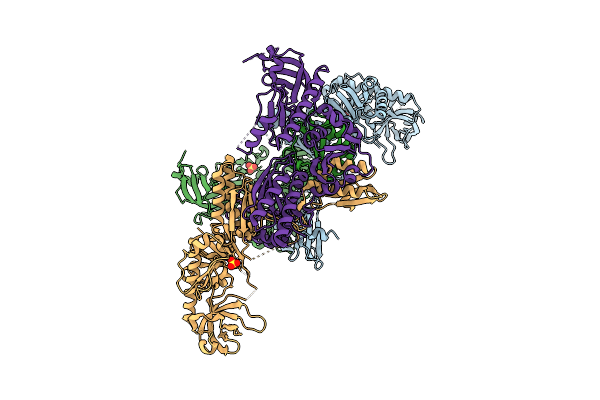 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-07-22 Classification: HYDROLASE Ligands: SO4, ZN |
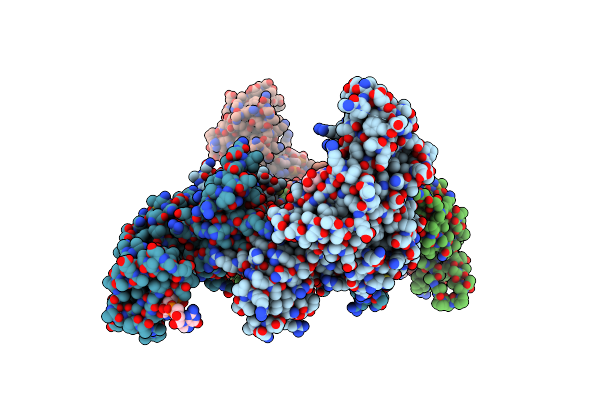 |
Crystal Structure Of E. Coli Rnase E Possessing M1 Rna Fragments - Catalytic Domain
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2008-07-22 Classification: HYDROLASE Ligands: ZN |
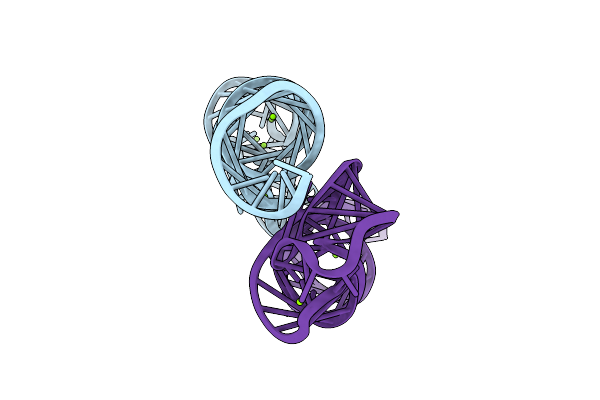 |
Method: X-RAY DIFFRACTION
Resolution:2.40 Å Release Date: 2008-07-15 Classification: RNA Ligands: MG |
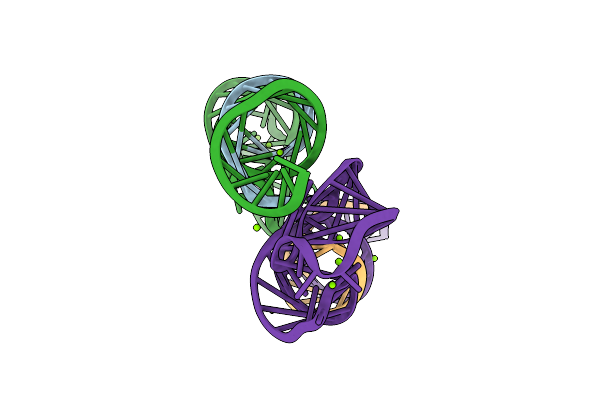 |
Method: X-RAY DIFFRACTION
Resolution:2.20 Å Release Date: 2008-07-15 Classification: RNA Ligands: MG |
 |
Method: X-RAY DIFFRACTION
Resolution:2.60 Å Release Date: 2007-03-13 Classification: RNA Ligands: MG |
 |
Method: X-RAY DIFFRACTION
Resolution:2.00 Å Release Date: 2007-02-20 Classification: RNA Ligands: MN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2005-10-25 Classification: HYDROLASE Ligands: MG, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2005-10-14 Classification: HYDROLASE Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2005-10-14 Classification: HYDROLASE Ligands: MG, ZN |
 |
The Structure Of A Rigorously Conserved Rna Element Within The Sars Virus Genome
Method: X-RAY DIFFRACTION
Resolution:2.70 Å Release Date: 2005-02-01 Classification: RNA Ligands: MG |
 |
Organism: Bradyrhizobium japonicum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-12-07 Classification: TRANSFERASE Ligands: HEM |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2004-02-24 Classification: RNA Ligands: CO, 5GP |

