Search Count: 22
 |
Structure Of Uncleaved Influenza A/Victoria/2570/2019 Haemagglutinin. The 2021 Influenza A(H1N1)Pdm09 Egg-Derived Vaccine Candidate.
Organism: H1n1 subtype
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2021-07-14 Classification: CELL CYCLE Ligands: PT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |
 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-12-18 Classification: HYDROLASE Ligands: MLI |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-03-07 Classification: Oxidoreductase/Chaperon Ligands: FAD, NA |
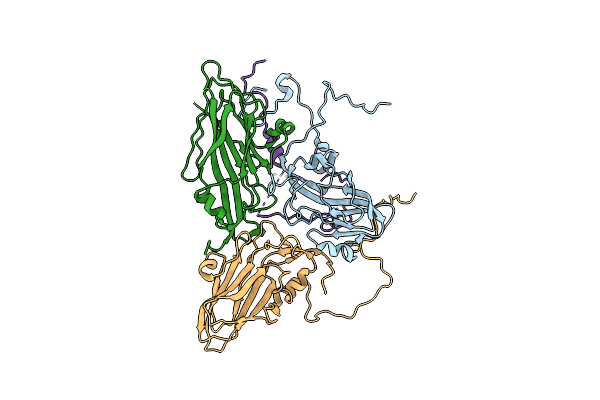 |
Structure-Based Energetics Of Protein Interfaces Guide Foot-And-Mouth Disease Virus Vaccine Design
Organism: Foot-and-mouth disease virus - type o
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2015-09-23 Classification: VIRUS |
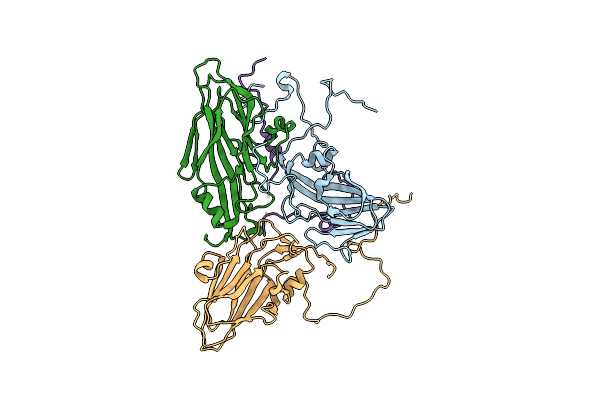 |
Structure-Based Energetics Of Protein Interfaces Guide Foot-And-Mouth Disease Virus Vaccine Design
Organism: Foot-and-mouth disease virus - type sat 2
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2015-09-23 Classification: VIRUS |
 |
Crystal Structure Of Recombinant Foot-And-Mouth-Disease Virus A22-H2093F Empty Capsid
Organism: Foot-and-mouth disease virus - type a
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-23 Classification: VIRUS |
 |
Crystal Structure Of Recombinant Foot-And-Mouth-Disease Virus O1M-S2093Y Empty Capsid
Organism: Foot-and-mouth disease virus - type o
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2015-09-23 Classification: VIRUS |
 |
Structure Of The Thermostable Alpha-Carbonic Anydrase From Thiomicrospira Crunogena Xcl-2 Gammaproteobacterium
Organism: Thiomicrospira crunogena (strain xcl-2)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-07-29 Classification: LYASE Ligands: ZN, BCT |
 |
Functionally Selective Inhibition Of Group Iia Phospholipase A2 Reveals A Role For Vimentin In Regulating Arachidonic Acid Metabolism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-17 Classification: HYDROLASE Ligands: CL, CA |
 |
Functionally Selective Inhibition Of Group Iia Phospholipase A2 Reveals A Role For Vimentin In Regulating Arachidonic Acid Metabolism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2012-10-17 Classification: HYDROLASE Ligands: U8D, CA, CL |
 |
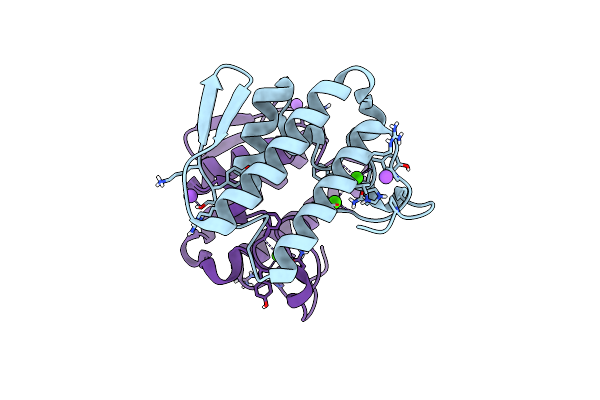
Functionally Selective Inhibition Of Group Iia Phospholipase A2 Reveals A Role For Vimentin In Regulating Arachidonic Acid Metabolism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-17 Classification: HYDROLASE Ligands: BHP, CA, CL |
 |
Functionally Selective Inhibition Of Group Iia Phospholipase A2 Reveals A Role For Vimentin In Regulating Arachidonic Acid Metabolism
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-10-17 Classification: HYDROLASE Ligands: CA, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-11-02 Classification: OXIDOREDUCTASE Ligands: ZN, FAD, AA |
 |
X-Ray Crystal Structure Of Nad(P)H: Quinone Oxidoreductase-1 (Nqo1) Bound To The Coumarin-Based Inhibitor As1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2010-01-12 Classification: OXIDOREDUCTASE Ligands: FAD, CC2 |
 |
Structural Basis Of N-End Rule Substrate Recognition In Escherichia Coli By The Clpap Adaptor Protein Clps
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-04-28 Classification: CHAPERONE |
 |
Structural Basis Of N-End Rule Substrate Recognition In Escherichia Coli By The Clpap Adaptor Protein Clps - The Phe Peptide Structure
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-04-28 Classification: PEPTIDE BINDING PROTEIN |
 |
Structural Basis Of N-End Rule Substrate Recognition In Escherichia Coli By The Clpap Adaptor Protein Clps - Trp Peptide Structure
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2009-04-28 Classification: PEPTIDE BINDING PROTEIN |

