Search Count: 39
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-01-24 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-15 Classification: METAL BINDING PROTEIN Ligands: AG, SO4, BTB |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-10-25 Classification: METAL BINDING PROTEIN Ligands: ZN |
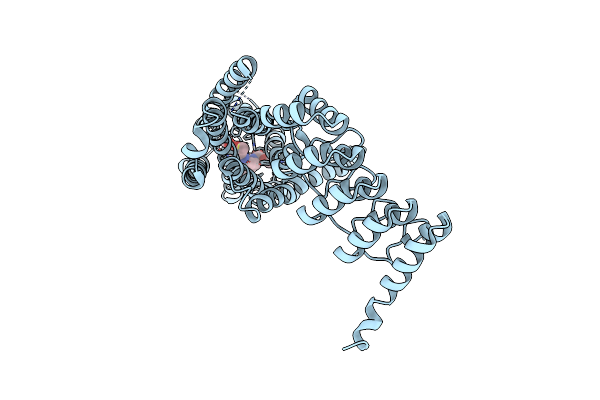 |
Crystal Structure Of The Human Alpha1B Adrenergic Receptor In Complex With Inverse Agonist (+)-Cyclazosin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2022-01-12 Classification: MEMBRANE PROTEIN Ligands: T0B |
 |
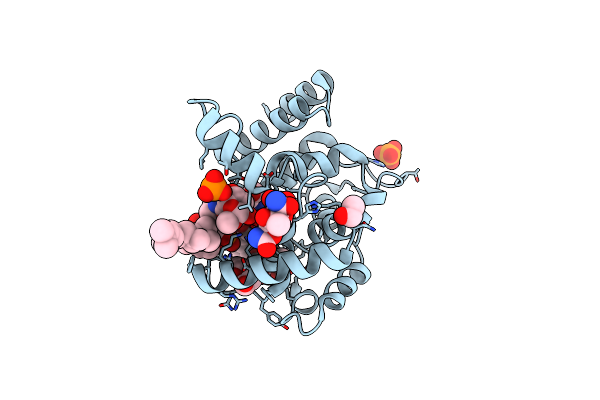
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, PO4, EDO |
 |
Crystal Structure Of S. Aureus Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, EDO, PEG, MG |
 |
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To A Seryl Sulfamoyl Adenosine Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: JPE, CL, SO4, DMS, EDO, TRS |
 |
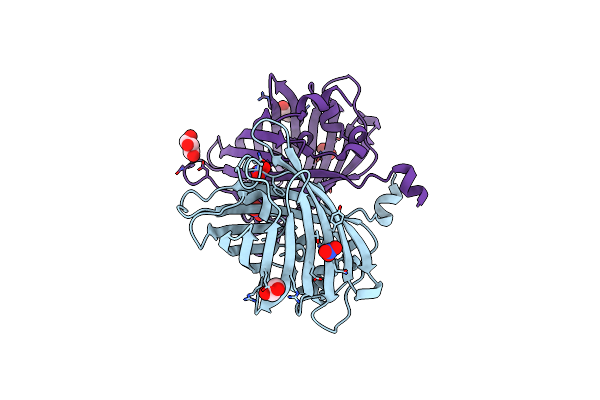
Staphylococcus Aureus Monofunctional Glycosyltransferase In Complex With Moenomycin
Organism: Staphylococcus aureus mw2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-06-27 Classification: TRANSFERASE Ligands: M0E, EDO, PO4, 1QW |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN Ligands: NO3, EDO, CL, GOL |
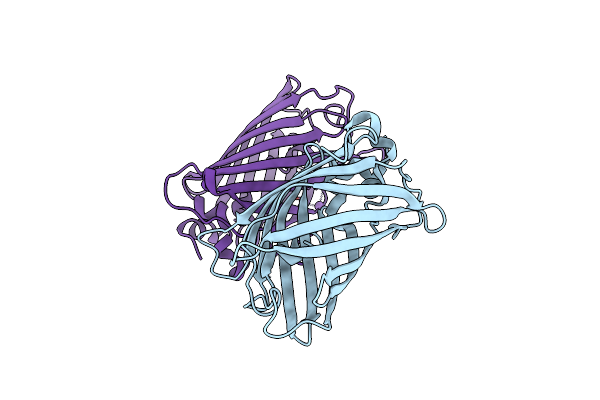 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-06 Classification: FLUORESCENT PROTEIN Ligands: CL, NA |
 |
Nmr Structure Of Kora, A Plasmid-Encoded, Global Transcription Regulator Kora
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2016-07-20 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Kora, A Plasmid-Encoded, Global Transcription Regulator
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-04-06 Classification: TRANSCRIPTION Ligands: ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-04-06 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Kora, A Plasmid-Encoded, Global Transcription Regulator
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-06 Classification: TRANSCRIPTION |
 |
Comparison Of The Structure And Activity Of Glycosylated And Aglycosylated Human Carboxylesterase 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-01-13 Classification: HYDROLASE Ligands: NAG, PO4 |
 |
Comparison Of The Structure And Activity Of Glycosylated And Aglycosylated Human Carboxylesterase 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2016-01-13 Classification: HYDROLASE Ligands: NAG |
 |
Comparison Of The Structure And Activity Of Glycosylated And Aglycosylated Human Carboxylesterase 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2016-01-13 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-05-07 Classification: HYDROLASE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-05-07 Classification: HYDROLASE |

