Search Count: 103
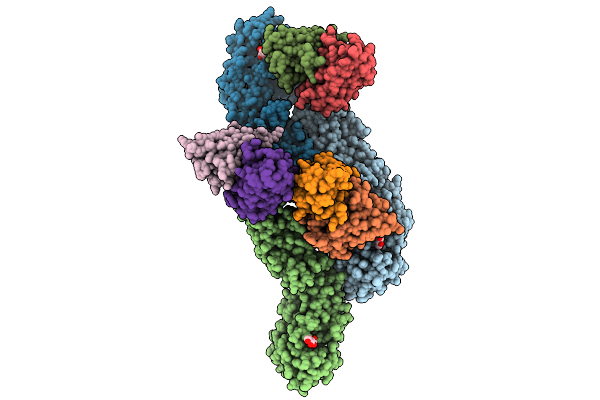 |
Organism: Yellow fever virus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRUS/IMMUNE SYSTEM Ligands: NAG |
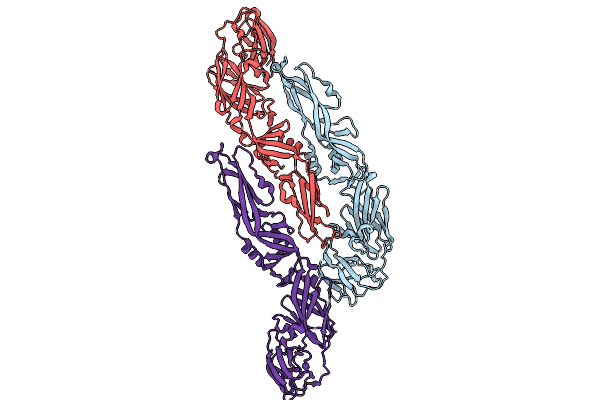 |
Organism: Yellow fever virus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRUS |
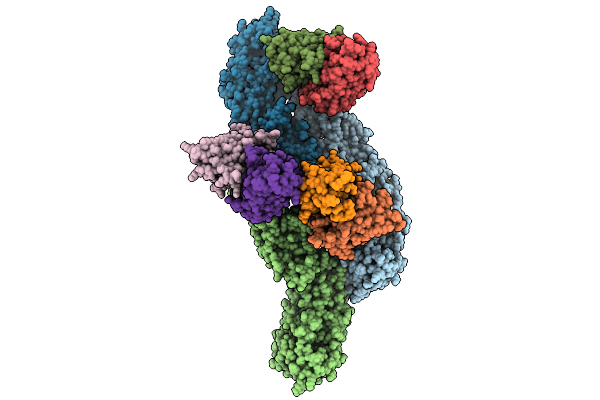 |
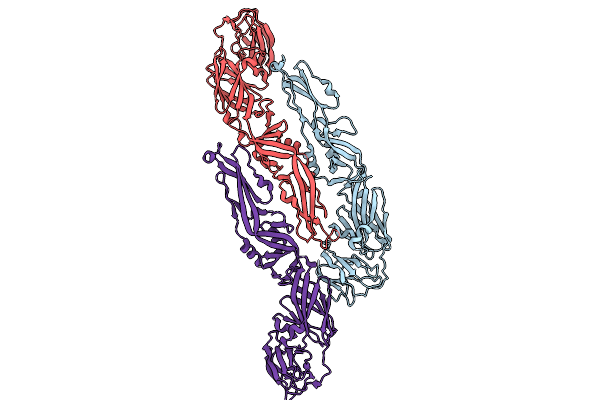
Envelope Protein Asu Of Yfv-Es504 With Yfv-17D Diii In Complex With 2C9 Fab
Organism: Yellow fever virus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRUS/IMMUNE SYSTEM |
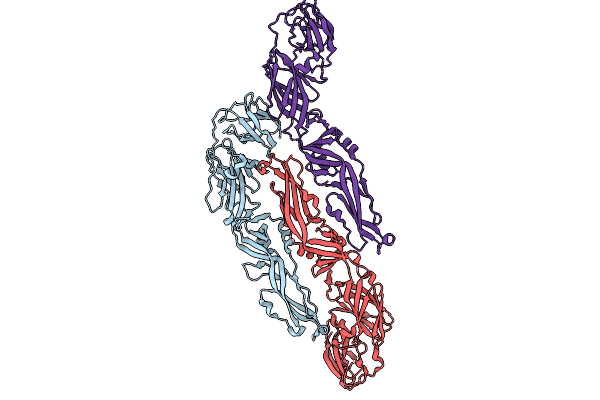 |
Organism: Yellow fever virus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRUS |
 |
Organism: Yellow fever virus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRUS |
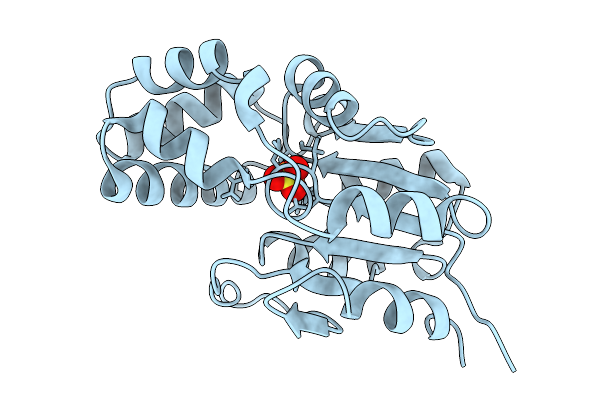 |
Crystal Structure Of Fluoroacetate Dehalogenase 4A From Delftia Acidovorans Strain D4B
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: SO4, CL |
 |
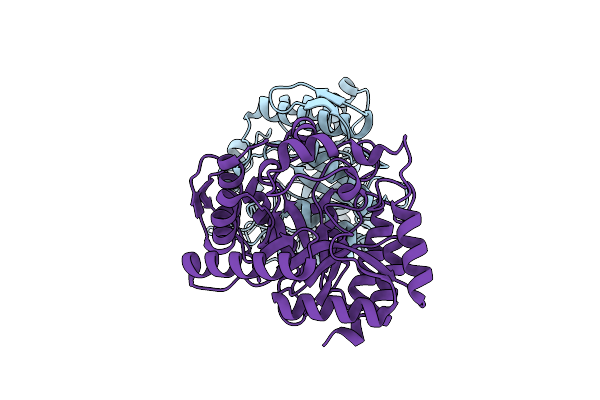
Crystal Structures Fluoroacetate Dehalogenase D4B From Delftia Acidovorans Strain D4B
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-02-05 Classification: FLAVOPROTEIN |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: PEG, NA |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL, NA, CL |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-01-01 Classification: HYDROLASE |
 |
Organism: Mycobacterium sp. js330
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-01-01 Classification: HYDROLASE Ligands: GOL, NA, CL, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: CL, SO4 |
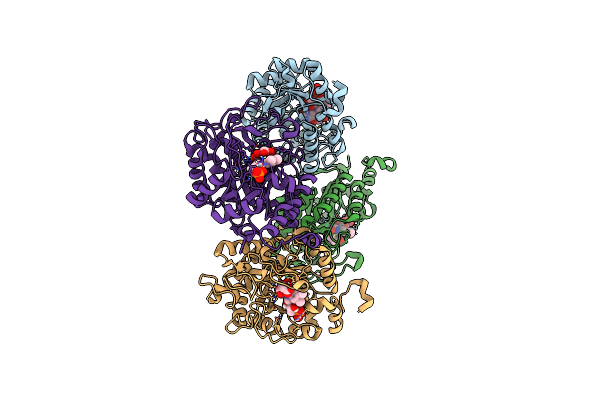 |
Succinate Bound Crystal Structure Of Thermus Scotoductus Sa-01 Ene-Reductase
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: SIN, FMN |
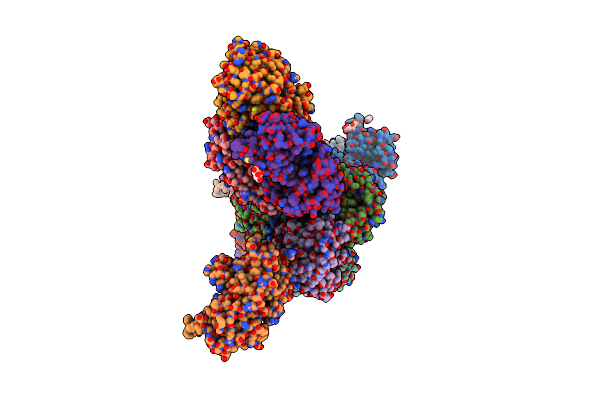 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 4B
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: ZN, W46, CIT, IPA |
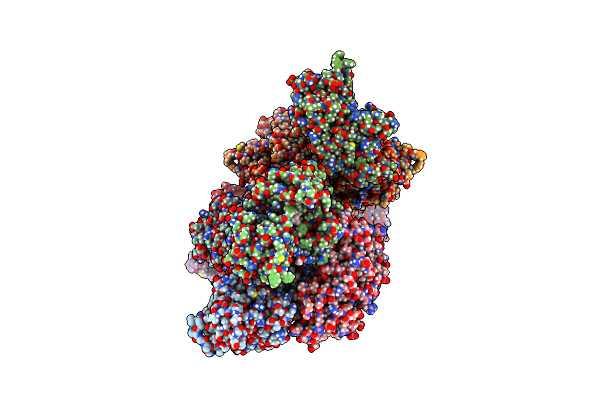 |
Rhodococcus Ruber Alcohol Dehydrogenase Nadh Biomimetic Complex - Compound 1A
Organism: Rhodococcus ruber
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: IPA, ZN, NA, W3O, CIT |
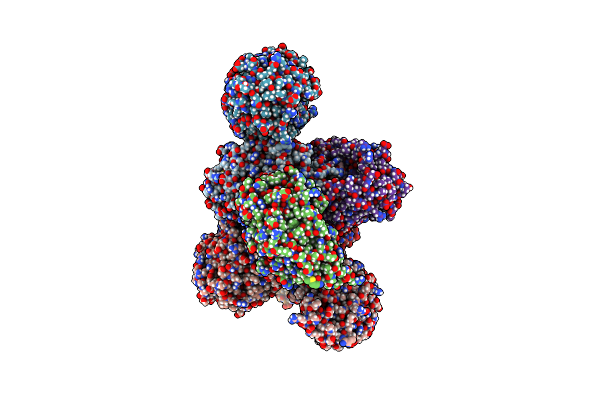 |
Organism: Thermus scotoductus sa-01
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-07-03 Classification: FLAVOPROTEIN Ligands: FMN, W3X, IPA, NA, CL |
 |
Organism: Methanococcoides burtonii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: EDO, CL, MG |
 |
Organism: Ramazzottius varieornatus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: TRANSFERASE Ligands: MEV, PEG, CA |
 |
Organism: Langya virus
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: NAG |

