Search Count: 86
 |
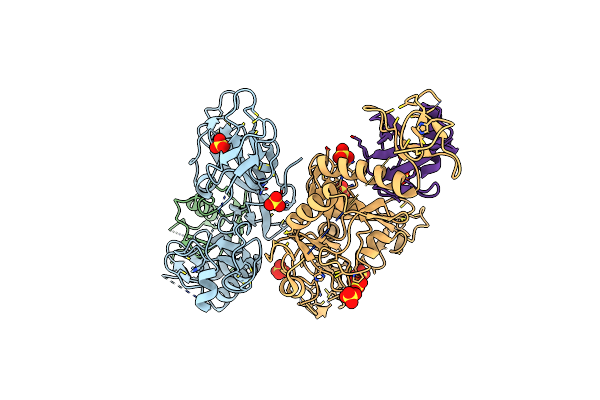
Organism: Synechocystis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: NI, GLY, 1PE, ACY |
 |
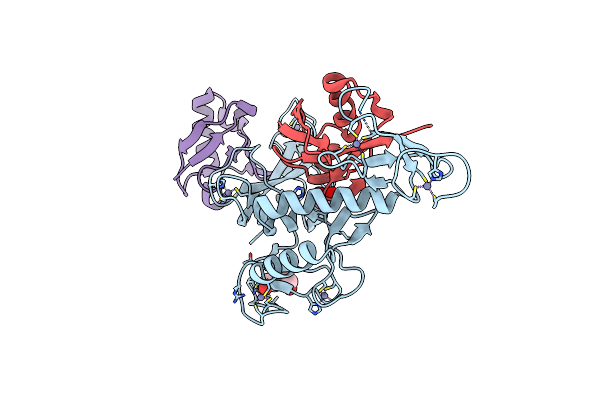
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-09-11 Classification: LIGASE Ligands: ZN, SO4 |
 |
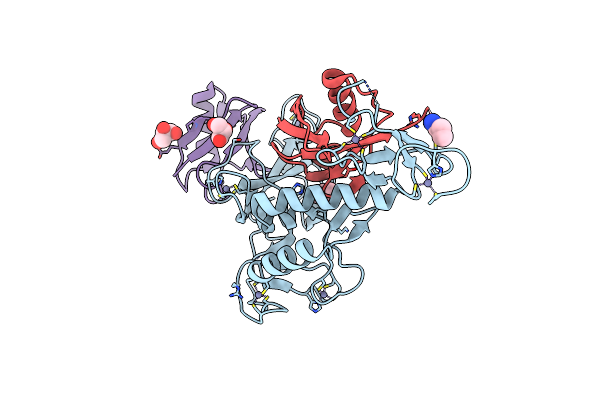
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-09-11 Classification: LIGASE Ligands: ZN, GOL, PEG |
 |
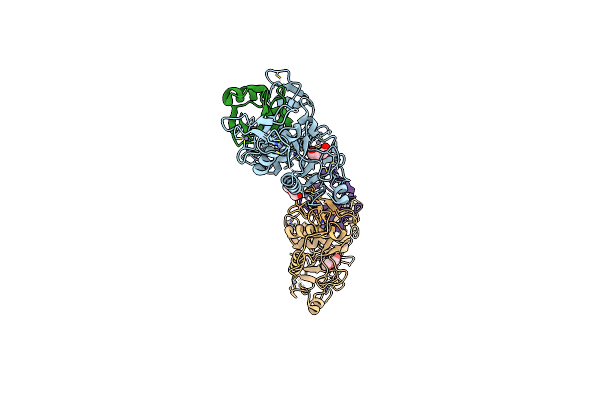
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-09-11 Classification: LIGASE Ligands: ZN, GOL, MPD, 3CN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-09-11 Classification: LIGASE Ligands: ZN, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-03 Classification: LIGASE Ligands: ZN, GOL, BA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: MG, GNP, GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: HYDROLASE Ligands: GDP, TVT |
 |
Organism: Lymantria dispar
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: GTP, MG |
 |
Organism: Lymantria dispar
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-11-08 Classification: VIRAL PROTEIN Ligands: GTP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION Ligands: U59, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION/INHIBITOR Ligands: U7R, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2023-03-29 Classification: GENE REGULATION/INHIBITOR Ligands: UHI, EDO |
 |
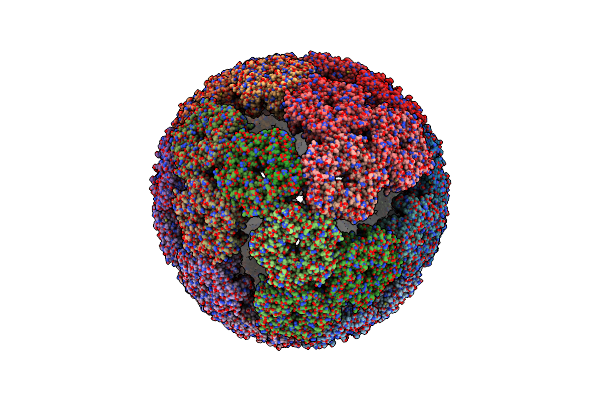
Aquifex Aeolicus Lumazine Synthase-Derived Nucleocapsid Variant Nc-1 (120-Mer)
Organism: Escherichia virus lambda, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRUS LIKE PARTICLE |
 |
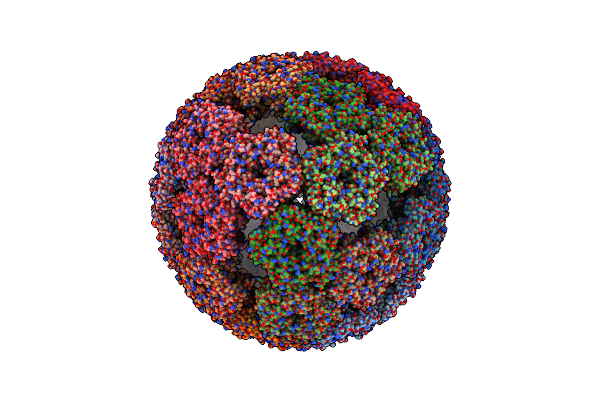
Aquifex Aeolicus Lumazine Synthase-Derived Nucleocapsid Variant Nc-1 (180-Mer)
Organism: Escherichia virus lambda, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRUS LIKE PARTICLE |
 |
Aquifex Aeolicus Lumazine Synthase-Derived Nucleocapsid Variant Nc-2 (180-Mer)
Organism: Escherichia virus lambda, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia virus lambda, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia virus lambda, Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: VIRUS LIKE PARTICLE |

