Search Count: 42
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Sinapic Acid, Tetragonal Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, SXX, SO4, NAG |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Sinapic Acid, Orthorhombic Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, SXX, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus Bound To Guaiacol, Orthorhombic Crystal
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4, JZ3 |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Reduced Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL, ACT |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, Oxidized Form
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, NA, CL, GOL |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, H166A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU1, GOL |
 |
Crystal Structure Of The Cupredoxin Acop From Acidithiobacillus Ferrooxidans, M171A Mutant
Organism: Acidithiobacillus ferrooxidans
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-09-13 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, GOL |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Ligand-Free Form
Organism: Pycnoporus cinnabarinus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Glucose-Bound Form
Organism: Pycnoporus cinnabarinus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, BGC, GLC, NAG, SO4 |
 |
Crystallographic Structure Of Oligosaccharide Dehydrogenase From Pycnoporus Cinnabarinus, Laminaribiose-Bound Form
Organism: Trametes cinnabarina
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-02-03 Classification: OXIDOREDUCTASE Ligands: FAD, GLC, NAG, SO4 |
 |
Olep, The Cytochrome P450 Epoxidase From Streptomyces Antibioticus Involved In Oleandomycin Biosynthesis: Functional Analysis And Crystallographic Structure In Complex With Clotrimazole.
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: HEM, CL6, SO4 |
 |
Organism: Lactococcus phage p2
Method: X-RAY DIFFRACTION Resolution:5.46 Å Release Date: 2014-07-09 Classification: VIRAL PROTEIN Ligands: CA |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: CA, MPG, TLA, C2G |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-14 Classification: TRANSFERASE Ligands: CA, MPG, SO4, C5P |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: TRANSFERASE Ligands: CA, CDP, MPG, TLA |
 |
Crystal Structure Of Urate Oxydase Using Surfactant Poloxamer 188 As A New Crystallizing Agent
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-02-23 Classification: OXIDOREDUCTASE Ligands: AZA, K |
 |
Structures Of Lactococcal Phage P2 Baseplate Shed Light On A Novel Mechanism Of Host Attachment And Activation In Siphoviridae
Organism: Lactococcus phage p2, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-02-16 Classification: VIRAL PROTEIN |
 |
Organism: Lactococcus phage p2
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2010-02-16 Classification: VIRAL PROTEIN Ligands: SR |
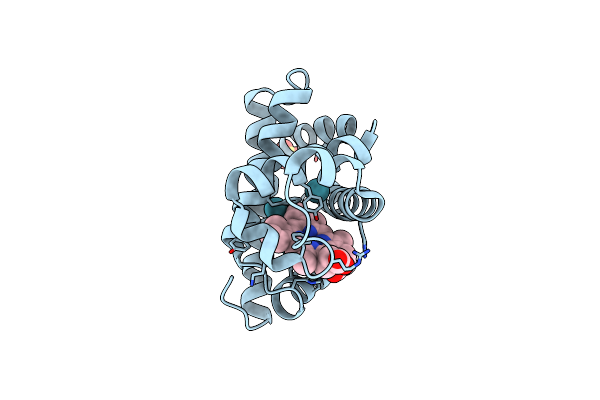 |
Crystal Structure Of Sperm Whale Myoglobin Mutant Yqrf In Complex With Xenon
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2009-12-22 Classification: OXYGEN STORAGE Ligands: HEM, XE, SO4 |
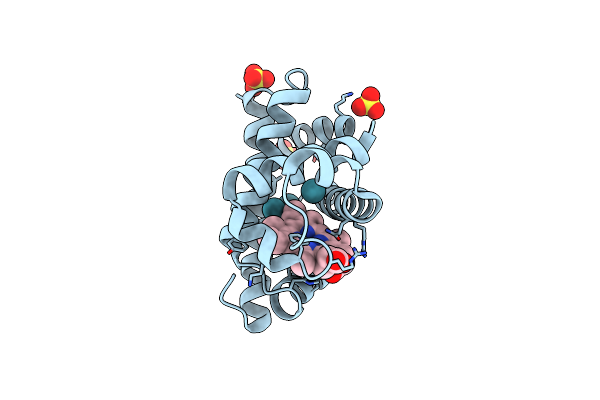 |
Crystal Structure Of Sperm Whale Myoglobin Mutant Yqr In Complex With Xenon
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-12-22 Classification: OXYGEN STORAGE Ligands: HEM, SO4, XE |

