Search Count: 25
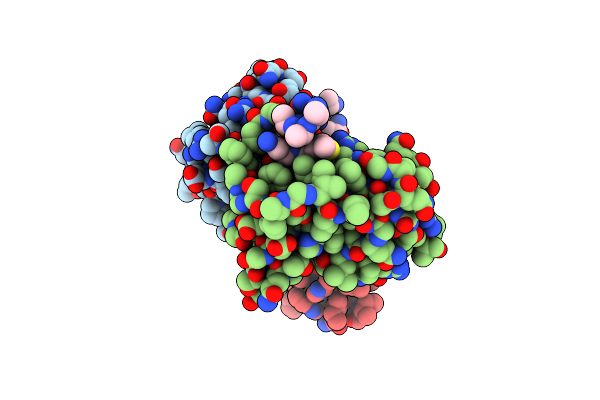 |
Cereblon Isoform 4 From Magnetospirillum Gryphiswaldense In Complex A Long Aspartimide Degron Peptide
Organism: Magnetospirillum gryphiswaldense, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-02-08 Classification: SIGNALING PROTEIN Ligands: ZN |
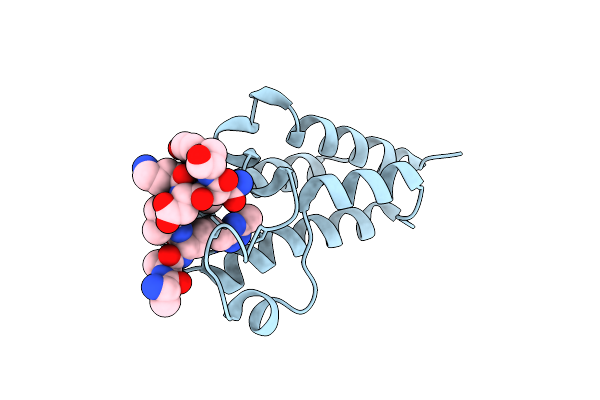 |
Human Brd3 Bromodomain 2 In Complex With A H4 Peptide Containing Apmtri (H4K20Apmtri)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-01-11 Classification: PEPTIDE BINDING PROTEIN |
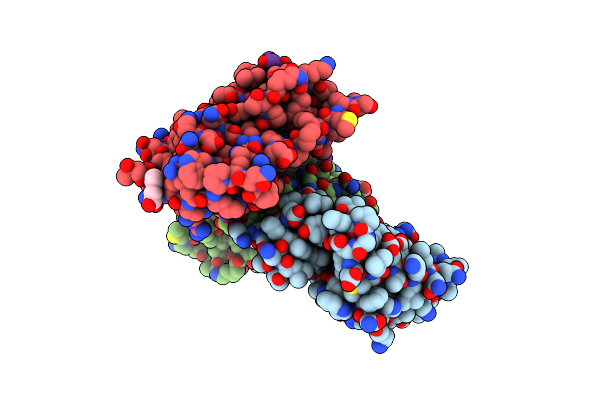 |
Human Brd4 Bromdomain 1 In Complex With A H4 Peptide Containing Acetyl Lysine And Apmtri (H4K5Ack8Apmtri)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-01-11 Classification: PEPTIDE BINDING PROTEIN Ligands: PEG |
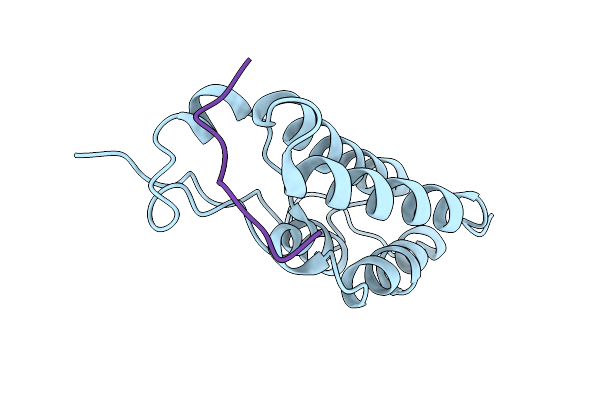 |
Human Brd4 Bromdomain 1 In Complex With A H4 Peptide Containing Apmtri (H4K5/8Apmtri)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-01-11 Classification: PEPTIDE BINDING PROTEIN |
 |
Cereblon Isoform 4 From Magnetospirillum Gryphiswaldense In Complex An Aspartimide Degron Peptide
Organism: Magnetospirillum gryphiswaldense, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-01-11 Classification: SIGNALING PROTEIN Ligands: ZN, PO4 |
 |
Cereblon Isoform 4 From Magnetospirillum Gryphiswaldense In Complex An Aminoglutarimide Degron Peptide
Organism: Magnetospirillum gryphiswaldense, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-01-11 Classification: SIGNALING PROTEIN Ligands: ZN, EF2 |
 |
Crystal Structure Of Arabidopsis Thaliana Naa60 In Complex With A Bisubstrate Analogue
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-06-24 Classification: PLANT PROTEIN Ligands: CMC |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-06-24 Classification: PLANT PROTEIN Ligands: ACO |
 |
Apo Crystal Structure Of The Colanidase Tailspike Protein Gp150 Of Phage Phi92
Organism: Enterobacteria phage phi92
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: SR, CL, GOL, EDO |
 |
Crystal Structure Of The Colanidase Tailspike Protein Gp150 Of Phage Phi92 Complexed With One Repeating Unit Of Colanic Acid
Organism: Enterobacteria phage phi92
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: SO4, MG, EDO, PG4 |
 |
Organism: Enterobacteria phage phi92
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-01-15 Classification: HYDROLASE,VIRAL PROTEIN Ligands: EDO, MG, CL, CA, NA |
 |
Crystal Structure Of The C-Terminal Fragment Of The Bacteriophage Phi92 Membrane-Piercing Protein Gp138
Organism: Bacteriophage phi92
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2012-02-22 Classification: VIRAL PROTEIN Ligands: FE, NA |
 |
Crystal Structure Of The Bacteriophage Phi92 Membrane-Piercing Protein Gp138
Organism: Bacteriophage phi92
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2012-02-22 Classification: VIRAL PROTEIN Ligands: FE, K |
 |
Crystal Structure Of The C-Terminal Fragment Of The Bacteriophage P2 Membrane-Piercing Protein Gpv
Organism: Enterobacteria phage p2
Method: X-RAY DIFFRACTION Resolution:0.94 Å Release Date: 2012-02-22 Classification: VIRAL PROTEIN Ligands: CL, CA, FE, NA |
 |
Organism: Enterobacteria phage p2
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2012-02-22 Classification: VIRAL PROTEIN Ligands: CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-15 Classification: TRANSCRIPTION Ligands: ZN |
 |
Crystal Structure Of Trim24 Phd-Bromo Complexed With H3(23-31)K27Ac Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-12-15 Classification: TRANSCRIPTION/Protein Binding Ligands: ZN |
 |
Crystal Structure Of Trim24 Phd-Bromo Complexed With H4(14-19)K16Ac Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-12-15 Classification: TRANSCRIPTION/Protein Binding Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-15 Classification: TRANSCRIPTION/Protein binding Ligands: ZN |
 |
Organism: Enterobacteria phage k1f
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2010-03-02 Classification: HYDROLASE |

