Search Count: 20
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-01-17 Classification: SIGNALING PROTEIN Ligands: PEG, MG, BEF |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-07 Classification: SIGNALING PROTEIN |
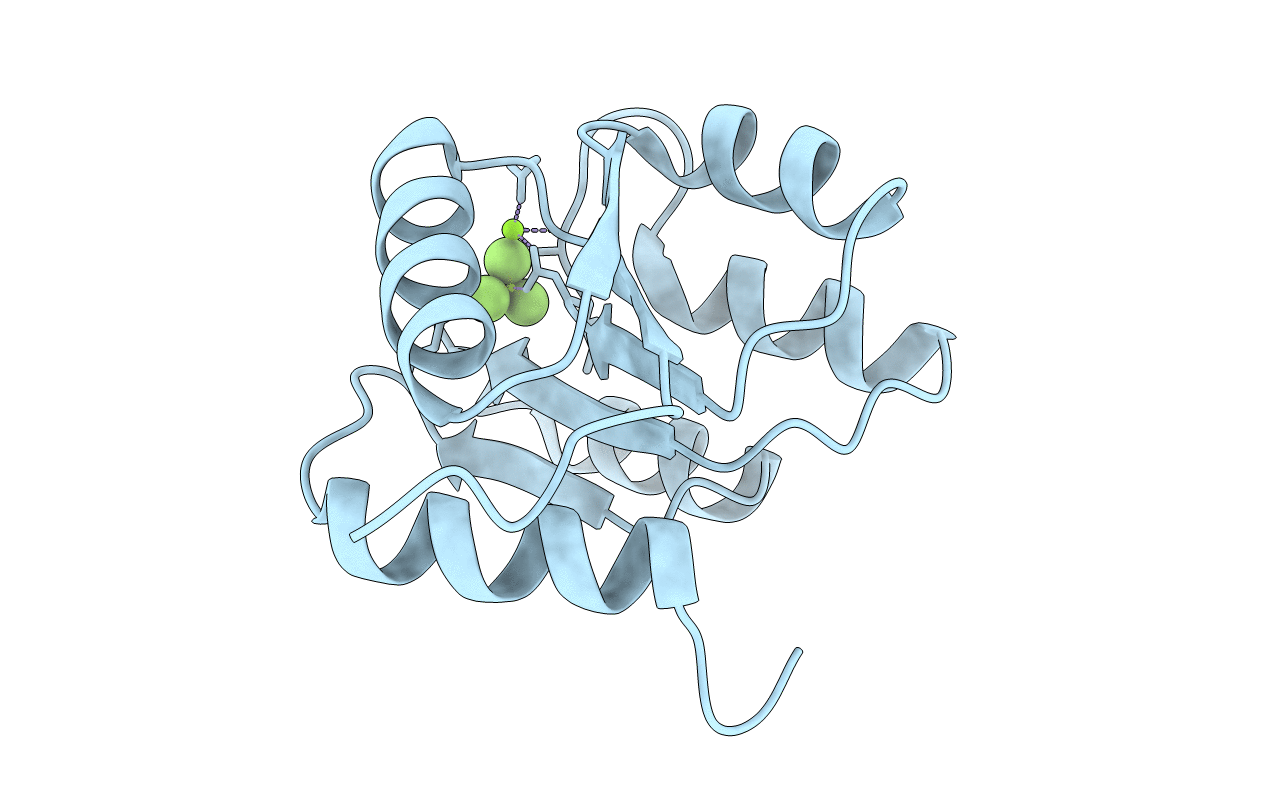 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-04-07 Classification: SIGNALING PROTEIN Ligands: MG, BEF |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2019-05-08 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-05-08 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With 1-Methyl-3-Indolyl-B-D-Galactopyranoside At 1.45 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, MHD, GAL |
 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With Chlorophenol Red-B-D-Galactopyranoside At 2.86 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, LRD, GAL |
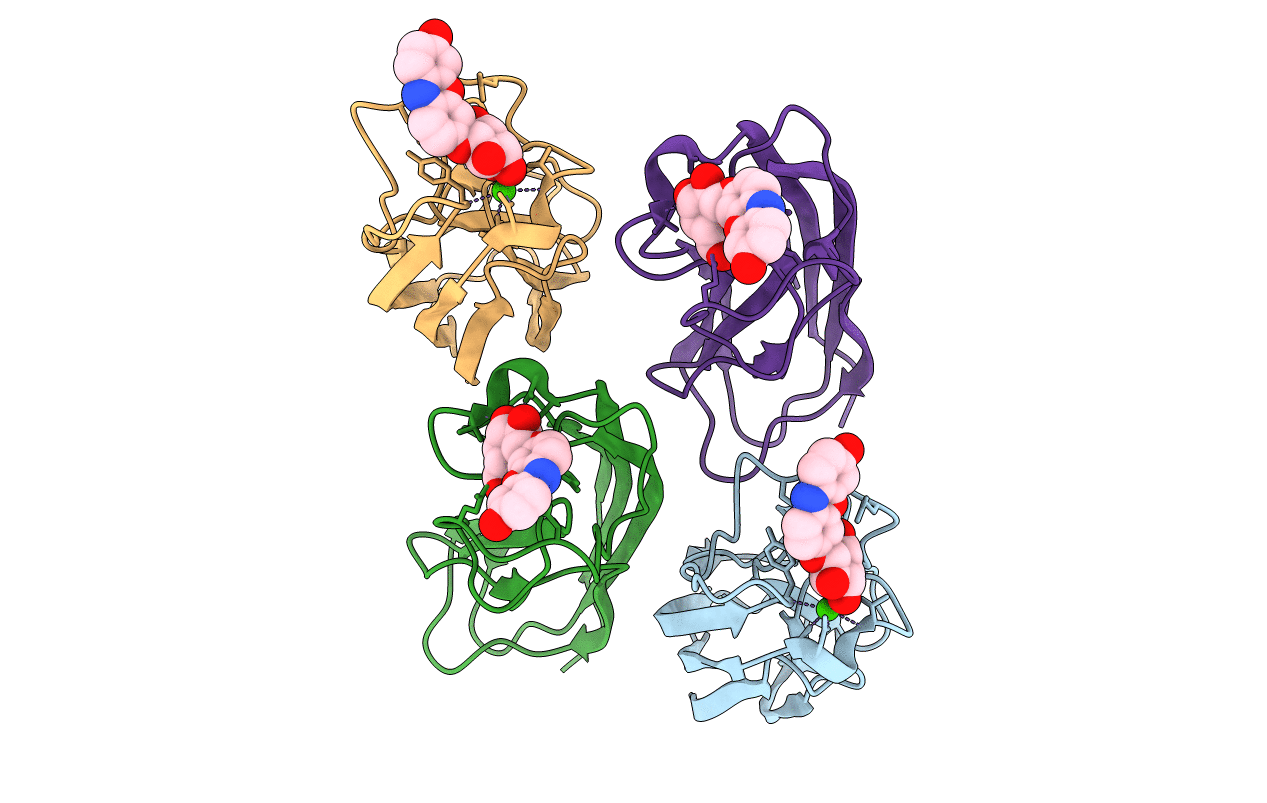 |
Crystal Structure Of Pseudomonas Aeruginosa Lectin Leca Complexed With Resorufin-B-D-Galactopyranoside At 1.76 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-10-30 Classification: SUGAR BINDING PROTEIN Ligands: CA, 04G, GAL |
 |
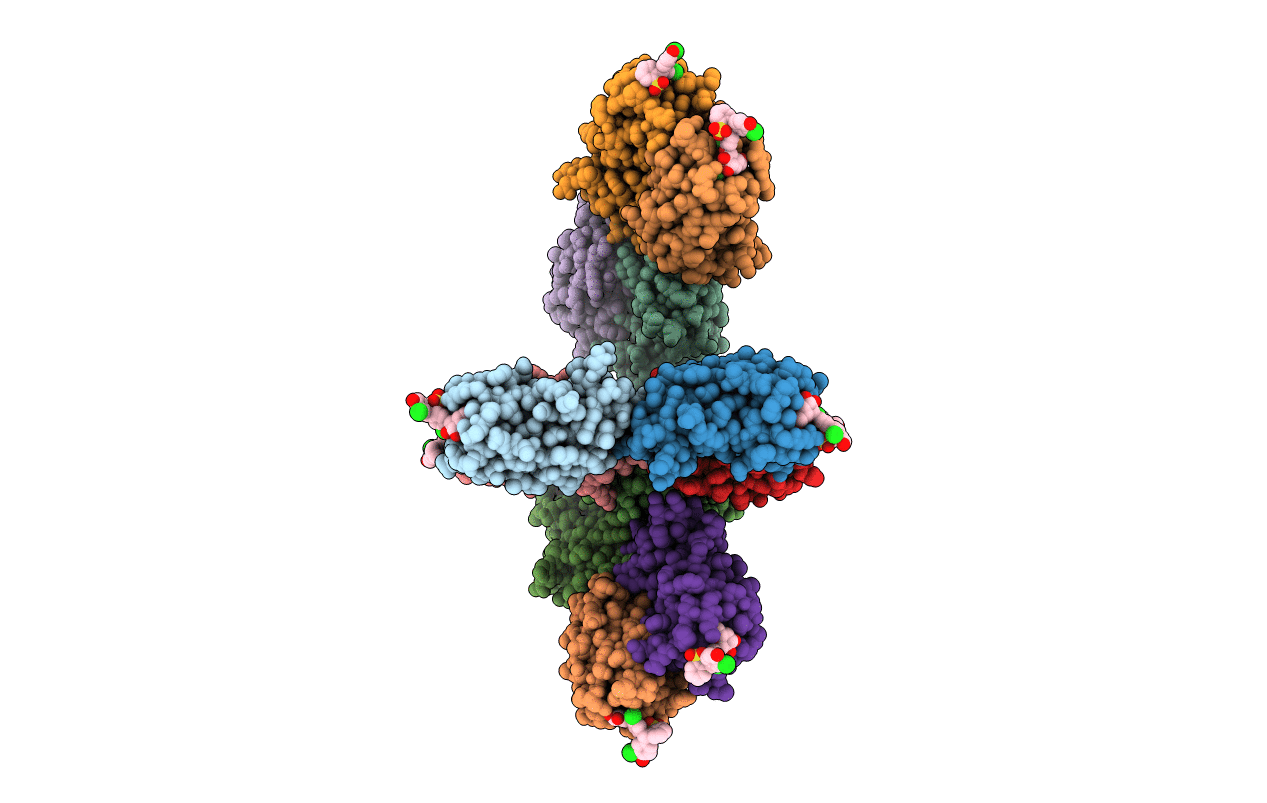
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-11-11 Classification: CELL CYCLE |
 |
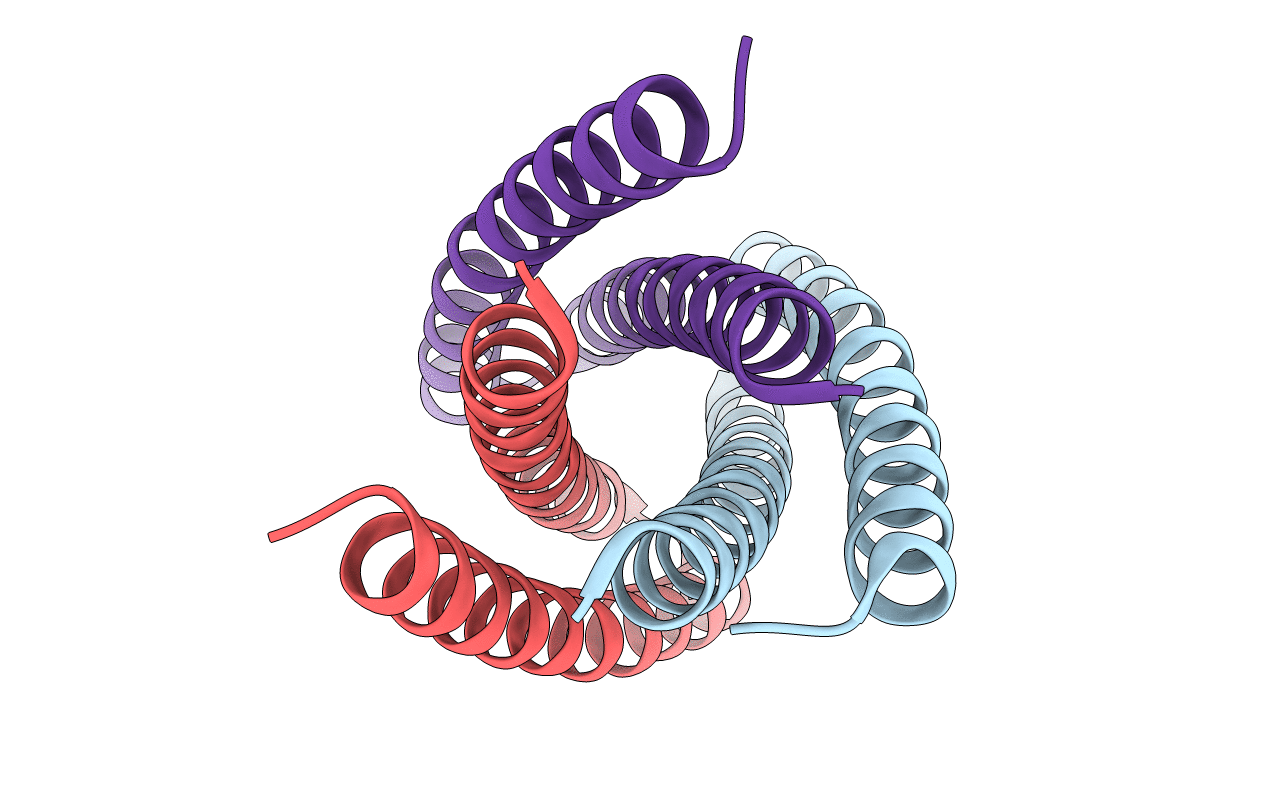
Midbody Targeting Of The Escrt Machinery By A Non-Canonical Coiled-Coil In Cep55
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-04 Classification: Cell cycle/Transport Protein |
 |
Structure Of Phosphorylated Enzyme I Of The Phosphoenolpyruvate:Sugar Phosphotransferase System
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: MG, OXL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-03-23 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-03-11 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2001-04-04 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-10-27 Classification: IMMUNE SYSTEM Ligands: CL, EDO |
 |
Structure Of Rat Farnesyl Protein Transferase Complexed With A Cvim Peptide And Alpha-Hydroxyfarnesylphosphonic Acid.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1999-06-18 Classification: TRANSFERASE Ligands: ACT, ZN, HFP |
 |
Organism: Human poliovirus 2
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 1998-09-16 Classification: VIRUS Ligands: SC4, MYR |
 |
Minor Conformer Of A Benzo[A]Pyrene Diol Epoxide Adduct Of Da In Duplex Dna
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 1998-08-19 Classification: DNA Ligands: BAP |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 1997-01-27 Classification: TOXIN |
 |
Structure Determination Of Antiviral Compound Sch 38057 Complexed With Human Rhinovirus 14
Organism: Human rhinovirus 14
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1993-10-31 Classification: VIRUS Ligands: S57 |

