Search Count: 13
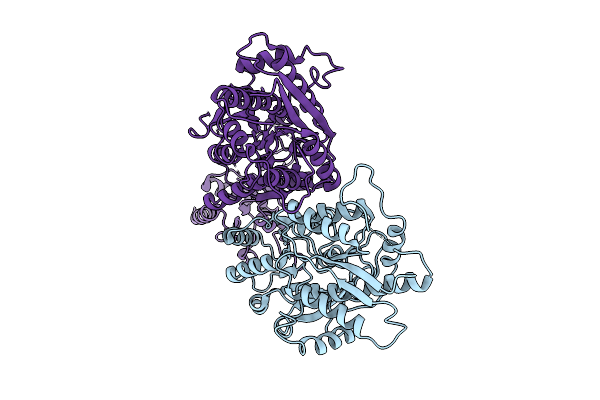 |
The Ntd Dimer And The Interfacing Lbd Region Of The Ampar Complex Glua3(R439G,R163I)- Tarp Gamma2 In The Apo State.
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN |
 |
The Tmd And The Lbd Region Of The Ampar Complex Glua3- Tarp Gamma2 In The Apo State.
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
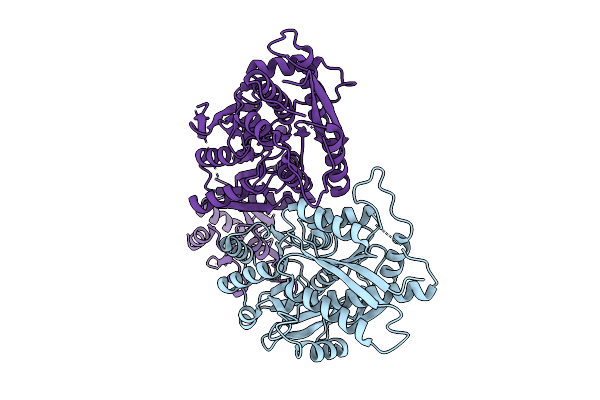 |
The Ntd Dimer And The Interfacing Lbd Region Of The Ampar Complex Glua3- Tarp Gamma2 In The Open State.
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
The Ntd Dimer And The Interfacing Lbd Region Of The Ampar Complex Glua3- Tarp Gamma2 In The Apo State
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: NAG |
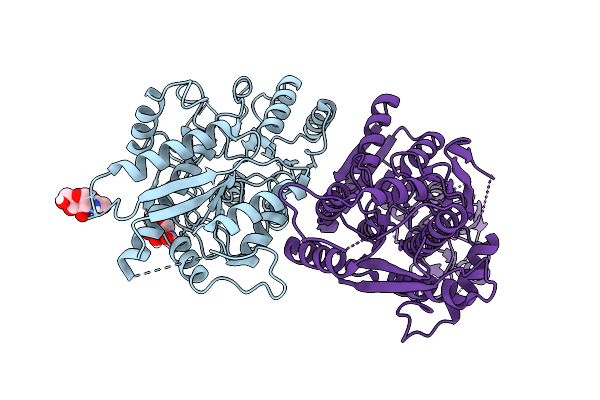 |
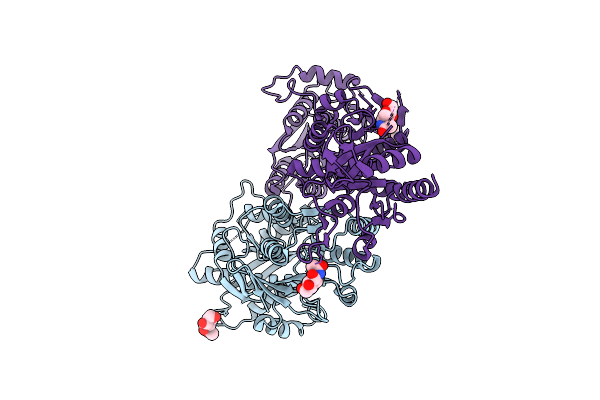
The Ntd Dimer And The Interfacing Lbd Region Of The Ampar Complex Glua3- Tarp Gamma2 In The Desensitised State.
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
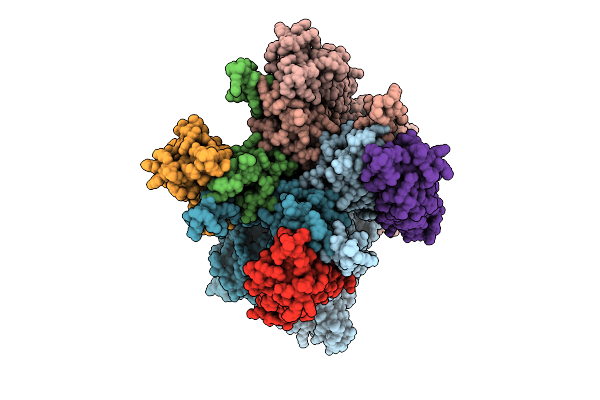
The Composite Map Of Of The Ampar Complex Glua3- Tarp Gamma2 In The Apo State.
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
 |
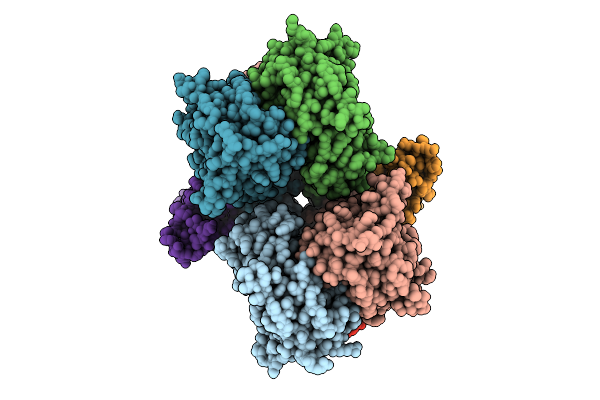
Cryo-Em Structure Of Cbf1-Ccan Bound Topologically To A Centromeric Cenp-A Nucleosome
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
 |
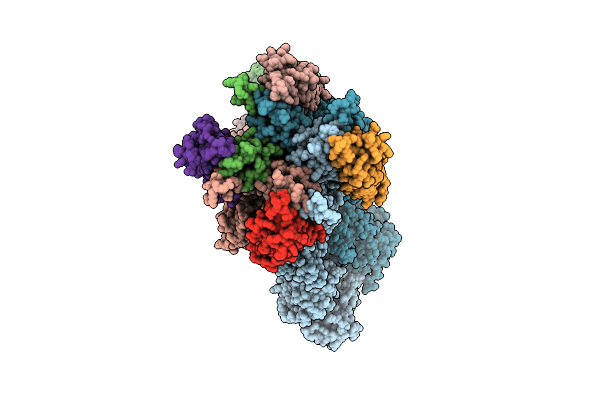
Cryo-Em Structure Of The Yeast Inner Kinetochore Bound To A Cenp-A Nucleosome.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: CELL CYCLE |
 |
E. Coli Beta-Hydroxydecanoyl Thiol Ester Dehydrase Modified By Its Classic Mechanism-Based Inactivator, 3-Decynoyl-N-Acetyl Cysteamine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-07-11 Classification: FATTY ACID BIOSYNTHESIS Ligands: DAC |
 |
Escherichia Coli Beta-Hydroxydecanoyl Thiol Ester Dehydrase At Ph 5 And 21 Degrees C
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-07-11 Classification: FATTY ACID BIOSYNTHESIS |

