Search Count: 31
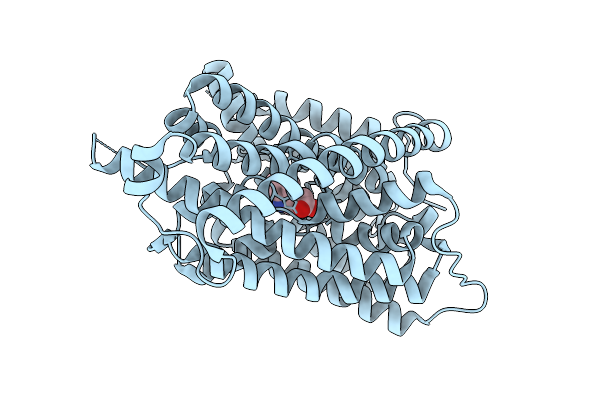 |
Auxin Transporter-Like Protein 3 (Lax3) In The Inward Occluded State In Complex With Auxin (Indole-3-Acetic Acid, Iaa)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: IAC |
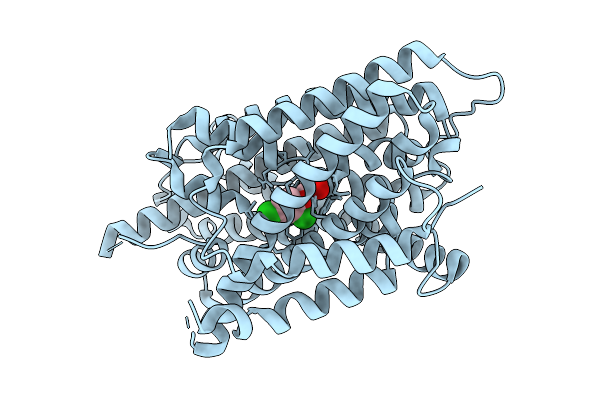 |
Auxin Transporter-Like Protein 3 (Lax3) In The Inward Occluded State In Complex With 2,4-Dichlorophenoxyacetic Acid (2,4-D)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: CFA |
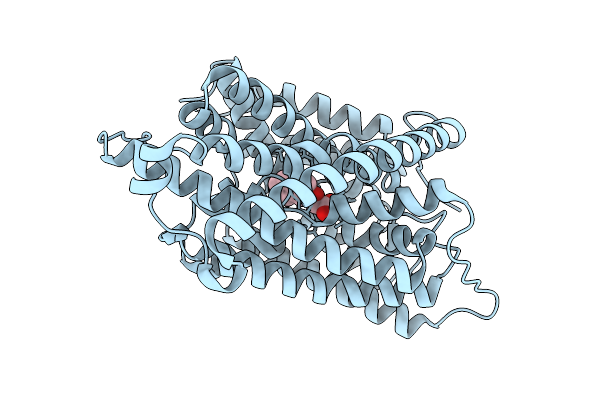 |
Auxin Transporter-Like Protein 3 (Lax3) In The Fully Occluded State In Complex With 2-Naphthoxyacetic Acid (2-Noa)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: A1ISN |
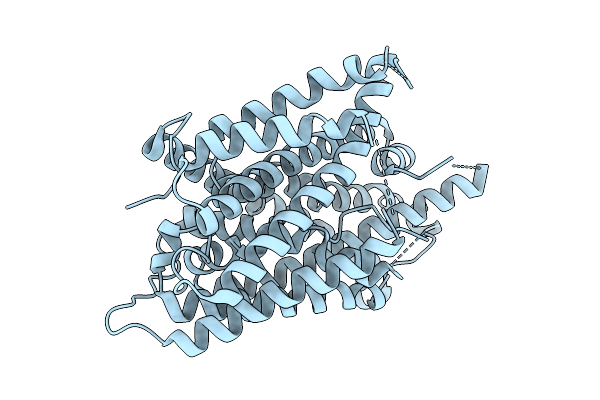 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN |
 |
Auxin Transporter Pin8 As Asymmetric Dimer (Inward/Outward) With 2,4-D Bound In The Inward Vestibule Prebinding State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: CFA, DMS, DLP |
 |
Auxin Transporter Pin8 As Asymmetric Dimer (Inward/Outward) With 4-Cpa Bound In The Inward Vestibule Prebinding State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: A1IHP, DLP |
 |
Auxin Transporter Pin8 As Symmetric Dimer (Outward/Outward) With 4-Cpa Bound In The Outward Binding State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: A1IHP, DLP |
 |
Auxin Transporter Pin8 As Asymmetric Dimer (Outward/Inward) With 4-Cpa Bound In The Outward Partly Released State
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: A1IHP, DMS, DLP |
 |
L2 Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Form I'' And Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-16 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex With Substitutions R269W, E271R, L273N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-02 Classification: LYASE Ligands: CAP, MG, B3P |
 |
Organism: Methylorubrum extorquens am1
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: LIGASE Ligands: COA, BTI |
 |
Organism: Methylorubrum extorquens am1
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: LIGASE Ligands: COA, BTI |
 |
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: S5Q, HCA, CLF, ADP, MG, AF3, SF4 |
 |
Chapso Treated Partial Catalytic Component (Comprising Only Anfd & Anfk, Lacking Anfg And Fefeco) Of Iron Nitrogenase From Rhodobacter Capsulatus
Organism: Rhodobacter capsulatus sb 1003
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: OXIDOREDUCTASE Ligands: CLF |
 |
L8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Form I'' And Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Ssu-Bearing Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8 Complex
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8 Complex With Substitution E170N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex With Substitution E170N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-12 Classification: LYASE Ligands: CAP, MG |

